Non-Targeted Authentication Approach for Extra Virgin Olive Oil
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

November 2016 NEMSN Newsletter
Vol. Vol. 26 No.No. 1 1 The National Eosinophilia-Myalgia Syndrome Network, Inc. November Page 2016 1 Friends Supporting Friends National EMS Network NEMSN National Eosinophilia-Myalgia Syndrome Network, Inc. http:www.nemsn.org Newsletter Points of interest ………………..……………... NEMSN is extremely pleased to announce that Stephen Naylor, • Your continued do- Ph.D. has joined our Medical Advisory Panel as NEMSN's nations have kept Science Advisor. Welcome, Dr. Naylor. this Newsletter and NEMSN alive. Dr. Stephen Naylor is one of only a handful of distin- Please keep those guished scientists who have devoted years of research in donations coming an attempt to achieve a complete explanation of EMS. no matter how Known as a chemist, biochemist, toxicologist and business large or small. Our originator, he has co-authored hundreds of scientific re- thanks. search papers and book chapters while delivering un- ………………………………….. counted presentations at universities and medical centers • We are interested worldwide. In the 1990s he and NEMSN advisor Gerald J. in your story. Please Gleich, M.D. partnered at the Mayo Clinic in Rochester, take the time to MN to undertake definitive EMS and L-tryptophan re- write it and send to search. Naylor and Gleich received NIH and WHO grants us for our newslet- and co-authored a number of scholarly papers during that period. ter. …………………………………... Stephen Naylor received his Ph.D. from the University of Cambridge (UK) in Biochem- • If you have not seen istry. He completed postdoctoral work at the Massachusetts Institute of Technology our web site yet, it where he later became a visiting faculty member in the Division of Biological Engineer- is very informative. -
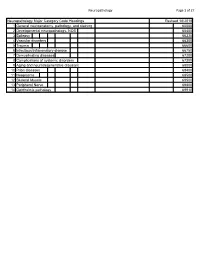
Neuropathology Category Code List
Neuropathology Page 1 of 27 Neuropathology Major Category Code Headings Revised 10/2018 1 General neuroanatomy, pathology, and staining 65000 2 Developmental neuropathology, NOS 65400 3 Epilepsy 66230 4 Vascular disorders 66300 5 Trauma 66600 6 Infectious/inflammatory disease 66750 7 Demyelinating diseases 67200 8 Complications of systemic disorders 67300 9 Aging and neurodegenerative diseases 68000 10 Prion diseases 68400 11 Neoplasms 68500 12 Skeletal Muscle 69500 13 Peripheral Nerve 69800 14 Ophthalmic pathology 69910 Neuropathology Page 2 of 27 Neuropathology 1 General neuroanatomy, pathology, and staining 65000 A Neuroanatomy, NOS 65010 1 Neocortex 65011 2 White matter 65012 3 Entorhinal cortex/hippocampus 65013 4 Deep (basal) nuclei 65014 5 Brain stem 65015 6 Cerebellum 65016 7 Spinal cord 65017 8 Pituitary 65018 9 Pineal 65019 10 Tracts 65020 11 Vascular supply 65021 12 Notochord 65022 B Cell types 65030 1 Neurons 65031 2 Astrocytes 65032 3 Oligodendroglia 65033 4 Ependyma 65034 5 Microglia and mononuclear cells 65035 6 Choroid plexus 65036 7 Meninges 65037 8 Blood vessels 65038 C Cerebrospinal fluid 65045 D Pathologic responses in neurons and axons 65050 1 Axonal degeneration/spheroid/reaction 65051 2 Central chromatolysis 65052 3 Tract degeneration 65053 4 Swollen/ballooned neurons 65054 5 Trans-synaptic neuronal degeneration 65055 6 Olivary hypertrophy 65056 7 Acute ischemic (hypoxic) cell change 65057 8 Apoptosis 65058 9 Protein aggregation 65059 10 Protein degradation/ubiquitin pathway 65060 E Neuronal nuclear inclusions 65100 -

Since January 2020 Elsevier Has Created a COVID-19 Resource Centre with Free Information in English and Mandarin on the Novel Coronavirus COVID- 19
Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company's public news and information website. Elsevier hereby grants permission to make all its COVID-19-related research that is available on the COVID-19 resource centre - including this research content - immediately available in PubMed Central and other publicly funded repositories, such as the WHO COVID database with rights for unrestricted research re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for free by Elsevier for as long as the COVID-19 resource centre remains active. Centers for Disease Control Stephen B Thackery, David J Sencery, and Harold W Jaffe, Centers for Disease Control and Prevention, Atlanta, GA, USA Ó 2017 Elsevier Inc. All rights reserved. This article is an updated version of the previous edition article by Stephen B. Thacker, David J. Sencer, volume 1, pp. 549–555, Ó 2008, Elsevier Inc. Early History of the Centers for Disease Control Health Officials and the American Public Health Association and Prevention agreed that CDC take the lead in defining what diseases were of highest priority and should be reportable. The key The history of the Centers for Disease Control and Prevention figure in this expansion of the CDC role was Alexander D. (CDC) began in 1942 with the establishment of the Malaria Langmuir, MD (1910–93), who was brought to the CDC in Control in War Areas (MCWA), under the U.S. -

Toxic Oil Syndrome
The WHO Regional OILSYNDROME TOXIC Offi ce for Europe The World Health Organization (WHO) is a specialized agency of the United Nations This is the third book published by WHO on the outbreak created in 1948 with the of the condition that came to be called the toxic oil primary responsibility for international syndrome (TOS), which struck Spain in 1981. It killed health matters and public health. The several hundred people and affected more than 20 000, WHO Regional Offi ce many of whom remain ill today. The two previous books for Europe is one of six regional offi ces described, respectively, early observations and scientifi c throughout the world, each with its own fi ndings gathered throughout the 1980s. programme geared to the particular health conditions of the This volume refl ects the progress made in the last ten countries it serves. years under a carefully planned strategy undertaken on Member States Albania | four main fronts by the WHO/CISAT Scientifi c Committee TENYEARS OF PROGRESS Andorra for the Toxic Oil Syndrome. First, various projects were Armenia supported aiming at the full chemical characterization Austria Azerbaijan of the oil matrix. Second, attempts were made to Belarus reproduce, on both a laboratory and an industrial scale, Belgium Bosnia and Herzegovina the refi ning process to which the suspect oil had been Bulgaria subjected in an attempt to establish the conditions Croatia Cyprus under which the toxin(s) were generated and to provide Czech Republic Denmark suffi cient amounts of reconstituted oils for toxicological Estonia studies. Third, a search was undertaken for an animal Finland France model in which to study the disease. -

Chapter 4 Prevention of Trichinella Infection in the Domestic
FAO/WHO/OIE Guidelines for the surveillance, management, prevention and control of trichinellosis Editors J. Dupouy-Camet & K.D. Murrell Published by: Food and Agriculture Organization of the United Nations (FAO) World Health Organization (WHO) World Organisation for Animal Health (OIE) The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations, of the World Health Organization and of the World Organisation for Animal Health concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The designations 'developed' and 'developing' economies are intended for statistical convenience and do not necessarily express a judgement about the stage reached by a particular country, territory or area in the development process. The views expressed herein are those of the authors and do not necessarily represent those of the Food and Agriculture Organization of the United Nations, of the World Health Organization and of the World Organisation for Animal Health. All the publications of the World Organisation for Animal Health (OIE) are protected by international copyright law. Extracts may be copied, reproduced, translated, adapted or published in journals, documents, books, electronic media and any other medium destined for the public, for information, educational or commercial purposes, provided prior written permission has been granted by the OIE. The views expressed in signed articles are solely the responsibility of the authors. The mention of specific companies or products of manufacturers, whether or not these have been patented, does not imply that these have been endorsed or recommended by FAO, WHO or OIE in preference to others of a similar nature that are not mentioned. -

Policy Responses to Reduce the Opportunity for Horsemeat Adulteration Fraud: the Case of the European Union
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by K-State Research Exchange POLICY RESPONSES TO REDUCE THE OPPORTUNITY FOR HORSEMEAT ADULTERATION FRAUD: THE CASE OF THE EUROPEAN UNION by MEGAN KULAS B.S., Kansas State University 2011 A THESIS submitted in partial fulfillment of the requirements for the degree MASTER OF SCIENCE Department of Diagnostic Medicine/Pathobiology College of Veterinary Medicine KANSAS STATE UNIVERSITY Manhattan, Kansas 2014 Approved by: Major Professor Justin Kastner Copyright MEGAN KULAS 2014 Abstract Food production is changing in response to an expanding global population. The ability to distribute and process ingredients amongst many individuals and countries has brought economic benefits while also creating new problems. By increasing the complexity of the supply chain, the food industry has birthed new dynamics, thus creating new opportunities for contamination, fraud, and other threats. One threat dynamic is the varying levels of food safety and quality control at different nodes along a supply chain. Contaminations pinpoint weaknesses of a supply chain, and such weaknesses could be exploited for harm. One way foods are intentionally contaminated is through food fraud. Food fraud involves substitution, mislabeling, dilution, and other means of criminal deception. Routine testing by an independent science- based group led to the discovery of one the largest scales of substitution and mislabeling in history—the 2013 adulteration of beef products with horsemeat. Commonly referred to as the horsemeat scandal of 2013, this important event in the history of the global food system affected several regions, hundreds of products, and thousands of retailers and consumers. -

WG Report Food and Environment
Working Group Food, Environment and Rare Diseases Summary The Working Group has involved a limited group of participants to ICORD 2010, possibly also because the topic of the role of dietary and environmental factors in rare diseases is rather new and still not clearly defined. On the other hand the discussion was lively and put the basis for further development of priority issues within ICORD activities. The following conclusions were derived and presented to the plenary ICORD meeting: − There is no doubt that quality and safety of food and environment are relevant to rare diseases (RD), hence, these have to be considered within ICORD mission and activities So, the relevant question is not whether , but how and in what instance. − In some instances, specific food- and environment-related factors are involved in the pathogenesis of RD. Telling, as well as widely recognized, examples are i) the insufficient intake of folates/folic acid concerning the pathogenesis of neural tube defects, representing the instance of a major risk factor; ii) aniline-denaturated rapeseed oil in toxic oil syndrome and C.Botulinum in infantile botulism, representing instances of primary causative agents. Likely, the progress of research on gene-environment interactions will increase the knowledge on specific risk factors involved in certain RD groups, such as birth defects and rare tumours. − In many other instances, diet or living environment may have an important modulating effect on the course, the severity, the prognosis of rare diseases. Telling examples, quoted during the discussion, were the high copper (and/or low zinc) in the diet for Wilson disease and inadequate intake of dietary lipids in Marfan disease. -

Hypereosinophilic Syndrome
In the light of recent advances: eosinophil, eosinophilia and idiopathic hypereosinophilic syndrome Aliflan YILDIRAN1, Aydan ‹K‹NC‹O⁄ULLARI1 1 Department of Pediatric Immunology and Allergy, Ankara University School of Medicine, Ankara, TURKEY Turk J Haematol 2005;22(3): 107-116 INTRODUCTION infiltration of multiple organs leading to se- vere organ dysfunction[3]. Recently, with the Eosinophil is a different cell containing discovery of different well-characterized un- highly toxic substances the functions of derlying molecular defects that ultimately le- which are still incompletely understood. Eo- ad to eosinophil expansion in some patients, sinophilia is a condition where the eosinophil the term “idiopathic” has become outdated in concentrations in the blood and some tissu- many cases[1]. es increase to unusual high levels in some abnormal conditions and disease states. Be- In the light of recent advances, we made ing a common finding in clinical practice, in an attempt to review the eosinophil, eosinop- the majority of cases, it can be ascribed to an hilia, IHES and the evaluation and manage- underlying disease[1]. The role of interleukin- ment of the latter. 5 (IL-5) in the induction of hypereosinophilia THE EOSINOPHIL in allergic diseases and parasitosis has been established recently[2]. It is an important task The eosinophil was first recognized 125 for clinicians to distinguish these conditions years ago as a distinct cellular element thro- from the very rare, but more serious idiopat- ugh the pioneering work of Paul Erlich. The hic hypereosinophilic syndrome (IHES), cha- ruddy dye, eosin, was named after the Greek racterized by persistent eosinophilia in perip- goddess of the morning sun, for use in histo- heral blood, bone marrow and eosinophilic logic staining by Erlich[4]. -

Dairy, Food and Environmental Sanitation 1990-07
ISSN: 1043-3546 July • 1990 90T8t? IW ‘dOa«W NNt» 502 E. Lincoln Way • Ames • Iowa • 5001 Vol • 10 • No. 7 • Pages 409-484 ayoy a332 maoN oui iyiM0IJ,':;'Na31NI swnidoaoiw Aiisa3AiNn ST/06 dX3 DAIRY, FOOD AND ENVIRONMENTAL SANITATION A Publication of the International Association of Milk, Food and Environmental Sanitarians, Inc. Please circle No. 176 on your Reader Service Card aiOUNM CHLORINE DIOXIDE PRODUCTS FOR INNOVATIVE PROBLEM SOLVING IN THE FOOD, MILK, DAIRY, AND BEVERAGE PROCESSING INDUSTRIES □ ENVIRONMENTAL CLEANERS SANITATION BEGINS WITH A GOOD ENVIRONMENTAL CLEANER CHLORINE DIOXIDE FOAM CHLORINE DIOXIDE CONVEYER LUBE CHLORINE DIOXIDE CIP SYSTEMS □ MICROBIOLOGICAL CONTROL IN PROCESS WATER COOLING TOWERS EFFLUENT TREATMENT POTABLE WATER TREATMENT □ RIO LINDA CHLORINE DIOXIDE PRODUCTS INTEGRATED SYSTEM ENGINEERING NEUTRAL EFFLUENT pH EPA REGISTERED PRECURSORS QUALIFIED TECHNICAL SUPPORT PROPRIETARY PATENTED TECHNOLOGY For Further Information Contact DON ENGLISH or LINDA HANNA RIO LINDA CHEMICAL 410 N. 10th STREET. SACRAMENTO, CA 95814 916-443-4939 800-822-5022 inside California or your local Rio Linda Representative stop by our Exhibit at the lAMFES Annuai Meeting BACTERIA CAN’T HIDE (LLNZADE® FROM P3-oxonia active® \ Service of ECOLAB^t) Peroxyacedc acid sanittzer. Please send: ■ S P □ Additional information about P3-oxoniaactive. :nvironmental sanitation program □ Literature about other Klenzade products that can help improve my plant's Environmental Sanitation Program. -or more information, complete and eturn this card, or call David Hurrry, NAME_ Dairy Plant Marketing Manager, at TITIF 612) 293-2525. COMPANY ADDRESS >top by our Exhibit at the fITV STAFF ZIP AMFES Annual Meeting □ Please phone me at: ( ) R<*sf tirnp- AM PM 01989 Ecolab Inc. -

Biopsy Diagnosis of Peripheral Neuropathy
Biopsy Diagnosis of Peripheral Neuropathy Juan M. Bilbao • Robert E. Schmidt Biopsy Diagnosis of Peripheral Neuropathy Second Edition Juan M. Bilbao, MD, FRCP (C) Robert E. Schmidt, MD, PhD Professor Emeritus of Neuropathology Professor of Pathology and Immunology Director Sunnybrook and St Michael’s Hospitals Division of Neuropathology University of Toronto Department of Pathology Toronto , ON Washington University School of Medicine Canada St. Louis , MO USA First published in 1995 by Butterworth-Heinemann, an imprint of Elsevier with the following title: Biopsy Diagnosis of Peripheral Neuropathy ISBN 978-3-319-07310-1 ISBN 978-3-319-07311-8 (eBook) DOI 10.1007/978-3-319-07311-8 Springer Cham Heidelberg New York Dordrecht London Library of Congress Control Number: 2014946574 © Springer International Publishing Switzerland 2015 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifi cally the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfi lms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. Exempted from this legal reservation are brief excerpts in connection with reviews or scholarly analysis or material supplied specifi cally for the purpose of being entered and executed on a computer system, for exclusive use by the purchaser of the work. Duplication of this publication or parts thereof is permitted only under the provisions of the Copyright Law of the Publisher's location, in its current version, and permission for use must always be obtained from Springer. -

Portable Through Bottle SORS for the Authentication of Extra Virgin Olive Oil
applied sciences Article Portable through Bottle SORS for the Authentication of Extra Virgin Olive Oil Mehrvash Varnasseri 1, Howbeer Muhamadali 1, Yun Xu 1, Paul I. C. Richardson 1, Nick Byrd 2, David I. Ellis 3 , Pavel Matousek 4 and Royston Goodacre 1,* 1 Department of Biochemistry and Systems Biology, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, BioSciences Building, Crown St., Liverpool L69 7ZB, UK; [email protected] (M.V.); [email protected] (H.M.); [email protected] (Y.X.); [email protected] (P.I.C.R.) 2 Campden BRI Group, Station Road, Chipping Campden GL55 6LD, UK; [email protected] 3 School of Chemistry, Manchester Institute of Biotechnology, University of Manchester, Manchester M1 7DN, UK; [email protected] 4 Central Laser Facility, Research Complex at Harwell, STFC Rutherford Appleton Laboratory, Harwell Oxford OX11 0QX, UK; [email protected] * Correspondence: [email protected] Abstract: The authenticity of olive oil has been a significant long-term challenge. Extra virgin olive oil (EVOO) is the most desirable of these products and commands a high price, thus unscrupulous individuals often alter its quality by adulteration with a lower grade oil. Most analytical methods employed for the detection of food adulteration require sample collection and transportation to a central laboratory for analysis. We explore the use of portable conventional Raman and spatially-offset Citation: Varnasseri, M.; Raman spectroscopy (SORS) technologies as non-destructive approaches to assess the adulteration Muhamadali, H.; Xu, Y.; Richardson, status of EVOO quantitatively and for SORS directly through the original container, which means P.I.C.; Byrd, N.; Ellis, D.I.; Matousek, that after analysis the bottle is intact and the oil would still be fit for use. -

Neurotoxicity?
Chapter 2 Introduction ‘‘Chemicals are an everyday fact of life in modern society. They enhance our lives in ways too numerous to count, but progress has its price, and too often the price of the role of chemicals in our society is human illness and disease. Representative Harold L. Volkmer Committee on Science and Technology U.S. House of Representatives October 8, 1985 ‘‘Nervous system dysfunction during advanced age seems destined to become the dominant disease entity of the twenty-first century. Neither I, nor anyone else, can tell you how much of that dysfunction might be attributable to toxic chemicals in the environment. So far, hardly anyone has looked. ” Bernard Weiss, Ph.D. Testimony before the Committee on Science and Technology U.S. House of Representatives October 8, 1985 CONTENTS Page WHAT IS NEUROTOXICITY? . ......’ ...*.*. ● .@ 43 SCOPE OF THIS STUDY . ....... ....... ....... ... 45 WHO IS AT RISK? . ..+..... ..+.*,*. ........ “. 45 EXAMPLES OF neurotoxic SUBSTANCE S. .. .+ .. .. ... ..+. 45 Industrial Chemicals . ........ ~.....+. ● ☛☛ 48 ,,..... Pesticides . ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ ✎ .....$+, -....... .......+ . 49 Therapeutic Drugs . 50 Abused Drugs . 51 Food Additives . 52 Cosmetics . 52 TOXIC SUBSTANCES AND NEUROLOGICAL AND PSYCHIATRIC DISORDERS . 54 IDENTIFYING neurotoxic SUBSTANCES . + . 55 REGULATING neurotoxic SUBSTANCES . 56 ECONOMIC CONSIDERATIONS . + . 56 INTERNATIONAL CONCERNS . + 56 CHAPTER 2 REFERENCES . + . + . 57 Page . 49 . .. 53 Page Fibers in the Cerebral Cortex . .. ,....52 the United States, . .. 55 in the United States, . .. 55 Page to Toxic Substances . 46 . .. 47 Chapter 2 Introduction Chemicals are an integral part of our daily lives number of neurotoxic substances in existence. The and are responsible for substantially improving number of neurotoxic substances that pose a them. Yet chemicals can also endanger our health, significant public health risk is unknown because even our survival.