Appendix a Software Environment for Plant Modeling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ted Nelson History of Computing
History of Computing Douglas R. Dechow Daniele C. Struppa Editors Intertwingled The Work and Influence of Ted Nelson History of Computing Founding Editor Martin Campbell-Kelly, University of Warwick, Coventry, UK Series Editor Gerard Alberts, University of Amsterdam, Amsterdam, The Netherlands Advisory Board Jack Copeland, University of Canterbury, Christchurch, New Zealand Ulf Hashagen, Deutsches Museum, Munich, Germany John V. Tucker, Swansea University, Swansea, UK Jeffrey R. Yost, University of Minnesota, Minneapolis, USA The History of Computing series publishes high-quality books which address the history of computing, with an emphasis on the ‘externalist’ view of this history, more accessible to a wider audience. The series examines content and history from four main quadrants: the history of relevant technologies, the history of the core science, the history of relevant business and economic developments, and the history of computing as it pertains to social history and societal developments. Titles can span a variety of product types, including but not exclusively, themed volumes, biographies, ‘profi le’ books (with brief biographies of a number of key people), expansions of workshop proceedings, general readers, scholarly expositions, titles used as ancillary textbooks, revivals and new editions of previous worthy titles. These books will appeal, varyingly, to academics and students in computer science, history, mathematics, business and technology studies. Some titles will also directly appeal to professionals and practitioners -
Ijiji '!!.=~:;~~~~"!:~~~!:T:I' LIFE Amer, Mosl Wanted Amel Mo.St Wanted Amel Most Wanled Amer
SA McCreary County Record, Tuessday, Apri124, 2012, Whitley City, Kentucky Saturday Evening April 28, 2012 .;11!1•• ;111•• 1'!'.... 113'!1. I I .111!1· ... ·f·I'II._t'..IICII- WATE/ABC The Blind Side Local TV GUIDE WVLTA:lSS NCIS The Mentalist 48 Hours Mystery Local WBIRINBC Escape Routes The Firm Law & Order: SVU Local Saturday Night Live WDKYIF0X... NASCAR Racing Aleafraz New Girl ILocal April 25 - May 1 AIl,E Storage IStorage Storage IStorage Flipped Oft Flipping Bosto.n Storage IStorage A~e Braveheart Braveheart CMT Road House Texas Women Southern Nights Texas Women Southern Nights CNN. CNN Presents Piers Mo.rgan To.night CNN Newsro.om CNN Presents 24/7 Mayweather e , , II ;; .. , , , , II II DISC Secret Service Killing bin Laden Seal Team 6 Auction Auction Seal Team 6 'MorJ F~m IApt. 23 Ravenga Loc;)I Nlgl:illl~a Jimmy Kimmel uve DISN Austin [Josslu Phineas IAustin Austin Austin Austin Jessie Jessie [Josslu IIfIIl'r1Oa5 Sul'liwr: One Wo~d CrimI 00I Min d~ :CSt C,lme 800110 Local: la10 Sh Ow Loll.IIl'IIm 'llalG WSIRINBC 8<lny Ie.FF E! Legally Blonde Kate·Will lce-Coc« The Soup Chelsea Fashio.n Po.lice WOK V/FOX Am eilc9.n idOl local ESPN NBA Basketball NBA BasKetball ESPN2 Ouarterback Year/Ouarterback Baseball Tonight SportsCenter SportsCenter SI..-a gil ISlora (/Cl lSI ora go IDug DuC!<D. Duel( D. DUel( D, IDuCKD, Siurage 'ISlorag1l FAM />JIIC Norm Counlry Pirates·Dea d Alice in Wonderland Finding Neverland CMf E> treme-Homs 'Ilea ventura At;(! Ventura FoX Dear John Dear John l.oule ILo.uie CNN Anderson Coiltler %0 Pllirs Morgan Tonlgl,t AndGf~O" Coiltler 360 E. -

SCMS 2019 Conference Program
CELEBRATING SIXTY YEARS SCMS 1959-2019 SCMSCONFERENCE 2019PROGRAM Sheraton Grand Seattle MARCH 13–17 Letter from the President Dear 2019 Conference Attendees, This year marks the 60th anniversary of the Society for Cinema and Media Studies. Formed in 1959, the first national meeting of what was then called the Society of Cinematologists was held at the New York University Faculty Club in April 1960. The two-day national meeting consisted of a business meeting where they discussed their hope to have a journal; a panel on sources, with a discussion of “off-beat films” and the problem of renters returning mutilated copies of Battleship Potemkin; and a luncheon, including Erwin Panofsky, Parker Tyler, Dwight MacDonald and Siegfried Kracauer among the 29 people present. What a start! The Society has grown tremendously since that first meeting. We changed our name to the Society for Cinema Studies in 1969, and then added Media to become SCMS in 2002. From 29 people at the first meeting, we now have approximately 3000 members in 38 nations. The conference has 423 panels, roundtables and workshops and 23 seminars across five-days. In 1960, total expenses for the society were listed as $71.32. Now, they are over $800,000 annually. And our journal, first established in 1961, then renamed Cinema Journal in 1966, was renamed again in October 2018 to become JCMS: The Journal of Cinema and Media Studies. This conference shows the range and breadth of what is now considered “cinematology,” with panels and awards on diverse topics that encompass game studies, podcasts, animation, reality TV, sports media, contemporary film, and early cinema; and approaches that include affect studies, eco-criticism, archival research, critical race studies, and queer theory, among others. -

Hartford Public Library DVD Title List
Hartford Public Library DVD Title List # 20 Wild Westerns: Marshals & Gunman 2 Days in the Valley (2 Discs) 2 Family Movies: Family Time: Adventures 24 Season 1 (7 Discs) of Gallant Bess & The Pied Piper of 24 Season 2 (7 Discs) Hamelin 24 Season 3 (7 Discs) 3:10 to Yuma 24 Season 4 (7 Discs) 30 Minutes or Less 24 Season 5 (7 Discs) 300 24 Season 6 (7 Discs) 3-Way 24 Season 7 (6 Discs) 4 Cult Horror Movies (2 Discs) 24 Season 8 (6 Discs) 4 Film Favorites: The Matrix Collection- 24: Redemption 2 Discs (4 Discs) 27 Dresses 4 Movies With Soul 40 Year Old Virgin, The 400 Years of the Telescope 50 Icons of Comedy 5 Action Movies 150 Cartoon Classics (4 Discs) 5 Great Movies Rated G 1917 5th Wave, The 1961 U.S. Figure Skating Championships 6 Family Movies (2 Discs) 8 Family Movies (2 Discs) A 8 Mile A.I. Artificial Intelligence (2 Discs) 10 Bible Stories for the Whole Family A.R.C.H.I.E. 10 Minute Solution: Pilates Abandon 10 Movie Adventure Pack (2 Discs) Abduction 10,000 BC About Schmidt 102 Minutes That Changed America Abraham Lincoln Vampire Hunter 10th Kingdom, The (3 Discs) Absolute Power 11:14 Accountant, The 12 Angry Men Act of Valor 12 Years a Slave Action Films (2 Discs) 13 Ghosts of Scooby-Doo, The: The Action Pack Volume 6 complete series (2 Discs) Addams Family, The 13 Hours Adventure of Sherlock Holmes’ Smarter 13 Towns of Huron County, The: A 150 Year Brother, The Heritage Adventures in Babysitting 16 Blocks Adventures in Zambezia 17th Annual Lane Automotive Car Show Adventures of Dally & Spanky 2005 Adventures of Elmo in Grouchland, The 20 Movie Star Films Adventures of Huck Finn, The Hartford Public Library DVD Title List Adventures of Ichabod and Mr. -
Marching for Healthy Babies
WEEKEND EDITION FRIDAY, APRIL 13 & SATURDAY, APRIL 14, 2012 | YOUR COMMUNITY NEWSPAPER SINCE 1874 | 75¢ Lake City Reporter LAKECITYREPORTER.COM Hair- raising efforts Loveloud A Wellborn-based United Way’s alternative rock/group, local impact Loveloud is the final per- formance in this season’s totals $1,075,973. FGC Entertainment series. The group, most recently By TONY BRITT seen on the Warped Tour, [email protected] perform on April 14th at Florida Gateway College. For more information or Few, if any men, want to for tickets, call (386) 754- lose most of their hair at 4340 or visit www.fgcenter- one time. tainment.com. But if losing his hair, in a public meeting, with a Toxic roundup pair of pink hair clippers, in front of close to 100 people The Columbia County meant the United Way of Toxic Roundup will be Suwannee Valley met its Saturday, April 14 at challenge goal, then Mike the Columbia County McKee, United Way presi- Fairgrounds from 9 a.m. dent took the buzz cut as a to 3 p.m. Safely dispose of COURTESY PHOTO good sport. your household hazard- Kaylie Spradley at 8 and a half months old in a photo taken Jan. 12. United Way of Suwannee ous wastes, including old Valley held its 2012 Royal paint, used oil, pesticides Awards Banquet and annual and insecticides. The meeting Thursday night at process is quick, easy Florida Gateway College and free of charge to Howard Conference Center, residents. There is a small Marching for chronicling the organiza- fee for businesses. Help keep our environment HAIR continued on 7A safe! For information call Columbia County landfill Healthy Babies at (386)752-6050. -

Ivan's Final Dissertation
Copyright by Ivan Angus Wolfe 2009 The Dissertation Committee for Ivan Angus Wolfe certifies that this is the approved version of the following dissertation: Arguing In Utopia: Edward Bellamy, Nineteenth Century Utopian Fiction, and American Rhetorical Culture Committee: Jeffrey Walker, Supervisor Mark Longaker Martin Kevorkian Trish Roberts-Miller Janet Davis Gregory Clark Arguing In Utopia: Edward Bellamy, Nineteenth Century Utopian Fiction, and American Rhetorical Culture by Ivan Angus Wolfe, A.A.S.; B.A.; M.A. Dissertation Presented to the Faculty of the Graduate School of English The University of Texas at Austin in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas at Austin May 2009 Dedication To Alexandra Acknowledgements I would like to thank my entire dissertation committee for their invaluable feedback during every step of my writing process. Jeffrey Walker was an invaluable director, always making time for my questions and responding with feedback to my chapters in a timely manner. Martin Kevorkian, Mark Longaker, Janet Davis, and Trish Roberts-Miller all also gave excellent advice and made themselves available whenever I needed them. Gregory Clark not only provided the initial impetus for my interest in this area, but graciously joined the committee despite his numerous commitments. Most graduate students would feel blessed to have such a conscientious and careful committee. I would also like to thank my wife, Alexandra, for her help. She has likely read this dissertation more times than anyone but me. Elizabeth Cullingford also deserves special thanks for being an excellent English department chair and making me feel welcome my first semester at UT as her Teaching Assistant for E316K. -
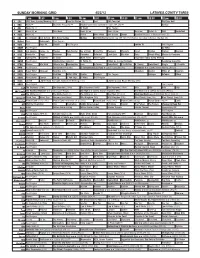
Sunday Morning Grid 4/22/12 Latimes.Com/Tv Times
SUNDAY MORNING GRID 4/22/12 LATIMES.COM/TV TIMES 7 am 7:30 8 am 8:30 9 am 9:30 10 am 10:30 11 am 11:30 12 pm 12:30 2 CBS CBS News Sunday Morning (N) Å Face the Nation (N) Paid PGA Tour Golf PGA Tour Golf 4 NBC News Å Meet the Press (N) Å Hockey Conference Quarterfinal: Teams TBA. (N) Å Hockey 5 CW News (N) Å In Touch Paid Program 7 ABC News (N) Å This Week News (N) Å News (N) Å News Å Vista L.A. NBA Basketball 9 KCAL News (N) Prince Mike Webb Joel Osteen Shook Baseball Dodgers at Houston Astros. (N) 11 FOX D. Jeremiah Joel Osteen Fox News Sunday Midday NASCAR Racing Sprint Cup: STP 400. From Kansas Speedway in Kansas City, Kan. (N) 13 MyNet Paid Tomorrow’s Paid Program 18 KSCI Paid Hope Hr. Church Paid Program Iranian TV Paid Program 22 KWHY Paid Program 22 Niños 24 KVCR Sid Science Curios -ity Thomas Bob Builder Joy of Paint Paint This Dewberry Wyland’s Sara’s Kitchen Kitchen Mexican 28 KCET Hands On Raggs Busytown Peep Pancakes Pufnstuf Land/Lost Hey Kids Taste Simply Ming Moyers & Company 30 ION Turning Pnt. Discovery In Touch Mark Jeske Paid Program Inspiration Today Camp Meeting 34 KMEX Paid Program Muchachitas Como Tú Al Punto (N) Fútbol de la Liga Mexicana República Deportiva 40 KTBN Rhema Win Walk Miracle-You Redemption Love In Touch PowerPoint It Is Written B. Conley From Heart King Is J. Franklin 46 KFTR Paid Program Toonturama Presenta El Campamento de Papá › (2007, Comedia) (PG) Blankman ›› (1994) Damon Wayans. -

Dream Machines Steven Connor a Lecture Given at the Object Emotions: Polemics Conference, University of Cambridge, 15Th April 2016
Dream Machines Steven Connor A lecture given at the Object Emotions: Polemics conference, University of Cambridge, 15th April 2016. The feelings we have for and in dreams are often mediated by the objects of which we dream (whether asleep or awake, and so taking dreaming in its largest sense), as well as the sorts of objects that dreams themselves may be taken to be. Those objects are sometimes, I think, mechanical in form and function. A machine is a thing. It is on the object side of things. Yet a machine is an anomalous kind of thing, an object that seems to exceed its objecthood in certain ways, through its quality of being automatic, of moving itself. Through its capacity for motion, a motor is an object that seems to be moving across into the condition of a subject, or quasi-subject. Machines do work for us, a machine is always a kind of substitute for a subject. And yet, as Michel Serres says, subjectivity is already substitution: ‘one must think of the subject as the potential for substitution. What does substitution mean? It is the same word as substance’ (Serres 2014, 88; my translation). A machine stands in vicariously for that which has, and so is the potential for vicariance itself. Subjects are not machines, because machines are objects; but they can imagine themselves as machines, as imaginary machines. A machine transmits force. It has motion rather than emotion. But what if the force transmitted by a machine is the force of fantasy, or what may come to the same thing, the fantasy of force? What kind of transport does such a machinery effect? The word transport moves between different registers of transmission – the physical movement of objects or energies and the movement of feeling, the feeling, for instance, of being, as we say, moved. -

Sunday Morning Grid 4/8/12
SUNDAY MORNING GRID 4/8/12 LATIMES.COM/TV TIMES 7 am 7:30 8 am 8:30 9 am 9:30 10 am 10:30 11 am 11:30 12 pm 12:30 2 CBS CBS News Sunday Morning (N) Å Face the Nation (N) Paid The 1987 Masters 2012 Masters Tournament Final Round. (N) Å 4 NBC News Å Meet the Press (N) Å Conference Wall Street Paid Program Willa’s Wild Pearlie Figure Skating 5 CW News (N) Å In Touch Paid Program Ann Shatilla 7 ABC News (N) Å This Week News (N) NBA Basketball Chicago Bulls at New York Knicks. (N) Å Golf Pre 9 KCAL News (N) Prince Mike Webb Joel Osteen Shook Paid Program 11 FOX D. Jeremiah Joel Osteen Fox News Sunday Midday Paid Program 13 MyNet Paid Tomorrow’s Paid Program Her Best Move (2007) (G) 18 KSCI Paid Hope Hr. Church Paid Program Iranian TV Paid Program 22 KWHY Paid Program Paid Program 24 KVCR Thomas & Friends Thomas Bob Builder Joy of Paint Paint This Dewberry Wyland’s Sara’s Kitchen Kitchen Mexican 28 KCET Hands On Raggs Busytown Peep Pancakes Pufnstuf Land/Lost Hey Kids Taste Simply Ming Moyers & Company 30 ION Turning Pnt. Discovery In Touch Mark Jeske Beyond Paid Program Inspiration Today Camp Meeting 34 KMEX Misa de Pascua (6) Muchachitas Como Tú Al Punto (N) Fútbol de la Liga Mexicana República Deportiva 40 KTBN Rhema Win Walk Miracle-You Redemption Love In Touch PowerPoint It Is Written B. Conley From Heart King Is J. Franklin 46 KFTR Paid Program Toonturama Presenta Nacida Para Ser Salvaje ›› (1995) Will Horneff. -

Felicity Walker (The Editor), at Felicity4711@ Gmail .Com Or #209–3851 Francis Road, Richmond, BC, Canada, V7C 1J6
The Newsletter of the British Columbia Science Fiction Association #482 $3.00/Issue July 2013 In This Issue: This and Next Month in BCSFA.....................................0 About BCSFA...............................................................0 Errata...........................................................................1 Letters of Comment......................................................1 Calendar......................................................................9 News-Like Matter.......................................................18 An Open Letter (Graeme Cameron)............................23 Book Launch Report: Part 1 (Joseph Picard)..............27 Zines Received...........................................................28 E-Zines Received........................................................29 Art Credits..................................................................30 BCSFAzine © July 2013, Volume 41, #7, Issue #482 is the monthly club newsletter published by the British Columbia Science Fiction Association, a social organiza- tion. ISSN 1490-6406. Please send comments, suggestions, and/or submissions to Felicity Walker (the editor), at felicity4711@ gmail .com or #209–3851 Francis Road, Richmond, BC, Canada, V7C 1J6. BCSFAzine solicits electronic submissions and black-and-white line illustrations in JPG, GIF, BMP, PNG, or PSD format, and offers printed contrib- utors’ copies as long as the club budget allows. BCSFAzine is distributed monthly at White Dwarf Books, 3715 West 10th Aven- ue, Vancouver, BC, -
Get Ready to Go out and Catch Some Fish/ Insert
$1 Midweek Edition Thursday, April 19, 2012 Reaching 110,000 Readers in Print and Online — www.chronline.com Scorched Woman, Child Escape Napavine Home Blaze / Main 16 Get Ready to Go Out and Catch Some Fish / Insert Missing on County Singer Mark Chesnutt Coming to Toledo / Main 9 the Chehalis Bearcat Boys, Girls Take Out Tigers Sheriff’s Office Continues Search for Olympia Man / Sports 1 Who Disappeared During the Pe Ell River Run By Kyle Spurr [email protected] Five days have passed since Daniel Kuhn was last seen in a small rubber raft floating down the Chehalis River near the Elk Creek Road Bridge Satur- day afternoon. Kuhn, dressed in a red hat, white tank top, blue jean shorts and white shoes, fell behind the rest of his group during the Pe Ell River Run and said he would meet them downriver near the Chandler Road Bridge, according to the Lewis County Sheriff’s Office. When Kuhn, 24, did not show up by Former Pe Ell Coach 8 p.m. Saturday, the rest of his group on Trial for Alleged Rape / Main 3 left. They thought he may have left with Lewis County Sheriff’s Office / Courtesy photo other friends, the sheriff’s office said. Daniel Kuhn, a 24-year-old Olympia man, By Monday, Kuhn’s friends realized was rafting with several other people on he was missing and contacted Lewis Saturday when he disappeared on the Chehalis River, somewhere upstream from County deputies. this spot on the Chandler Road Bridge. The Chehalis Fire Department’s swift water rescue team searched a three-mile portion of the Chehalis River between Elk Creek Road and the Chandler Road Bridge Tuesday afternoon in kayaks for clues. -

Tv Pg 7 4-24.Indd
The Goodland Star-News / Tuesday, April 24, 2012 7 All Central Time, for Kansas Mountain TIme Stations subtract an hour TV Channel Guide Tuesday Evening April 24, 2012 7:00 7:30 8:00 8:30 9:00 9:30 10:00 10:30 11:00 11:30 35 NFL 67 Bravo 22 ESPN 41 Hallmark ABC Last Man Cougar Dancing With Stars Private Practice Local Nightline Jimmy Kimmel Live S&T Eagle 37 USA 68 truTV 23 ESPN 2 45 NFL CBS NCIS NCIS: Los Angeles Unforgettable Local Late Show Letterman Late 2 PBS KOOD 2 PBS KOOD 24 ESPN Nws 47 Food NBC The Biggest Loser The Voice Fashion Star Local Tonight Show w/Leno Late 38 TBS 71 SCI FI 3 KWGN WB 3 NBC-KUSA 25 TBS 49 E! FOX Glee New Girl New Girl Local 39 WGN 72 Spike 4 ABC-KLBY Cable Channels 5 KSCW WB 26 Animal 51 Travel A&E Storage Storage Storage Storage Storage Storage Storage Storage Wars Local 40 TNT 73 Comedy 6 Weather 27 VH1 54 MTV 6 ABC-KLBY AMC O Brother, Where Art Thou? O Brother, Where Art Kindergar Local 41 FX 74 MTV 7 CBS-KBSL 28 TNT 55 Discovery ANIM Frozen Planet Wild Russia Frozen Planet Local 7 KSAS FOX 8 NBC-KSNK 42 Discovery 75 VH1 29 CNBC 56 Fox Nws BET The Game The Game The Game Together The Game Together Wendy Williams Show Good Hair Local 8 NBC-KSNK 9 Eagle 30 FSN RM 57 Disney BRAVO Housewives/OC Housewives/OC Housewives/OC Local Housewives/OC Atlanta 43 TLC 76 CMT 9 NBC-KUSA 11 QVC 31 CMT 58 History CMT Local Local Young Guns Young Guns II Young Guns 44 HGTV 77 EWTN 12 CW2-KWGN CNN Piers Morgan Tonight Anderson Cooper 360 E.