Glycosyl-Phosphatidylinositol/Inositol Phosphoglycan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(4,5) Bisphosphate-Phospholipase C Resynthesis Cycle: Pitps Bridge the ER-PM GAP
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by UCL Discovery Topological organisation of the phosphatidylinositol (4,5) bisphosphate-phospholipase C resynthesis cycle: PITPs bridge the ER-PM GAP Shamshad Cockcroft and Padinjat Raghu* Dept. of Neuroscience, Physiology and Pharmacology, Division of Biosciences, University College London, London WC1E 6JJ, UK; *National Centre for Biological Sciences, TIFR-GKVK Campus, Bellary Road, Bangalore 560065, India Address correspondence to: Shamshad Cockcroft, University College London UK; Phone: 0044-20-7679-6259; Email: [email protected] Abstract Phospholipase C (PLC) is a receptor-regulated enzyme that hydrolyses phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) at the plasma membrane (PM) triggering three biochemical consequences, the generation of soluble inositol 1,4,5-trisphosphate (IP3), membrane– associated diacylglycerol (DG) and the consumption of plasma membrane PI(4,5)P2. Each of these three signals triggers multiple molecular processes impacting key cellular properties. The activation of PLC also triggers a sequence of biochemical reactions, collectively referred to as the PI(4,5)P2 cycle that culminates in the resynthesis of this lipid. The biochemical intermediates of this cycle and the enzymes that mediate these reactions are topologically distributed across two membrane compartments, the PM and the endoplasmic reticulum (ER). At the plasma membrane, the DG formed during PLC activation is rapidly converted to phosphatidic acid (PA) that needs to be transported to the ER where the machinery for its conversion into PI is localised. Conversely, PI from the ER needs to be rapidly transferred to the plasma membrane where it can be phosphorylated by lipid kinases to regenerate PI(4,5)P2. -

Non-Canonical Regulation of Phosphatidylserine Metabolism by a Phosphatidylinositol Transfer Protein and a Phosphatidylinositol 4-OH Kinase
bioRxiv preprint doi: https://doi.org/10.1101/696336; this version posted July 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Non-Canonical Regulation of Phosphatidylserine Metabolism by a Phosphatidylinositol Transfer Protein and a Phosphatidylinositol 4-OH Kinase Yaxi Wang1,2, Peihua Yuan2, Ashutosh Tripathi2, Martin Rodriguez1, Max Lönnfors2, Michal Eisenberg-Bord3, Maya Schuldiner3, and Vytas A. Bankaitis1,2,4† 1Department of Biochemistry and Biophysics Texas A&M University College Station, Texas 77843-2128 USA 2Department of Molecular and Cellular Medicine Texas A&M Health Science Center College Station, Texas 77843-1114 USA 3Department of Molecular Genetics Weizmann Institute of Science, Rehovot 7610001, Israel 4Department of Chemistry Texas A&M University College Station, Texas 77840 USA Key Words: phosphoinositides/ PITPs/ lipid kinases/ lipi metabolism/ membrane contact site † -- Corresponding author TEL: 979-436-0757 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/696336; this version posted July 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. ABSTRACT The phosphatidylserine (PtdSer) decarboxylase Psd2 is proposed to engage in an endoplasmic reticulum (ER)-Golgi/endosome membrane contact site (MCS) that facilitates phosphatidylserine decarboxylation to phosphatidylethanomaine (PtdEtn) in Saccharomyces cerevisiae. -
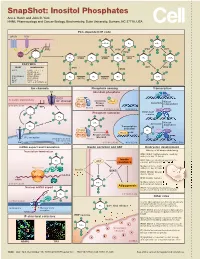
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

(4,5)-Bisphosphate Destabilizes the Membrane of Giant Unilamellar Vesicles
5112 Biophysical Journal Volume 96 June 2009 5112–5121 Profilin Interaction with Phosphatidylinositol (4,5)-Bisphosphate Destabilizes the Membrane of Giant Unilamellar Vesicles Kannan Krishnan,† Oliver Holub,‡ Enrico Gratton,‡ Andrew H. A. Clayton,§ Stephen Cody,§ and Pierre D. J. Moens†* †Centre for Bioactive Discovery in Health and Ageing, School of Science and Technology, University of New England, Armidale, Australia; ‡Laboratory for Fluorescence Dynamics, Department of Biomedical Engineering, University of California, Irvine, California; and §Ludwig Institute for Cancer Research, Royal Melbourne Hospital, Victoria, Australia ABSTRACT Profilin, a small cytoskeletal protein, and phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2] have been implicated in cellular events that alter the cell morphology, such as endocytosis, cell motility, and formation of the cleavage furrow during cytokinesis. Profilin has been shown to interact with PI(4,5)P2, but the role of this interaction is still poorly understood. Using giant unilamellar vesicles (GUVs) as a simple model of the cell membrane, we investigated the interaction between profilin and PI(4,5)P2. A number and brightness analysis demonstrated that in the absence of profilin, molar ratios of PI(4,5)P2 above 4% result in lipid demixing and cluster formations. Furthermore, adding profilin to GUVs made with 1% PI(4,5)P2 leads to the forma- tion of clusters of both profilin and PI(4,5)P2. However, due to the self-quenching of the dipyrrometheneboron difluoride-labeled PI(4,5)P2, we were unable to determine the size of these clusters. Finally, we show that the formation of these clusters results in the destabilization and deformation of the GUV membrane. -

GRAS Notice No. GRN 000728, FDA Has No Questions, Phospholipase C
.. .. Janet Oesterling Novozymes North America, Inc. 77 Perry Chapel Church Road Franklinton, NC 27525 Re: GRAS Notice No. GRN 000728 Dear Ms. Oesterling: The Food and Drug Administration (FDA, we) completed our evaluation of GRN 000728. We received Novozymes North America, Inc.’s (Novozymes’) notice on August 25, 2017, and filed it on September 8, 2017. We received an amendment containing additional safety information on October 11, 2017. The subject of the notice is phosphoinositide phospholipase C enzyme preparation produced by Bacillus licheniformis expressing a modified synthetic gene encoding a variant of the wild-type phosphoinositide phospholipase C from Pseudomonas sp. 62186 (phosphoinositide phospholipase C enzyme preparation) for use as an enzyme in the degumming of vegetable oils at a maximum level of 1.4 mg Total Organic Solids (TOS)/kg oil. The notice informs us of Novozymes’ view that this use of phosphoinositide phospholipase C enzyme preparation is GRAS through scientific procedures. Commercial enzyme preparations that are used in food processing typically contain an enzyme component that catalyzes the chemical reaction as well as substances used as stabilizers, preservatives, or diluents. Enzyme preparations may also contain components derived from the production organism and from the manufacturing process, e.g., constituents of the fermentation media or the residues of processing aids. Novozymes’ notice provides information about the components in the phosphoinositide phospholipase C enzyme preparation. Per the classification system of enzymes established by the International Union of Biochemistry and Molecular Biology, phosphoinositide phospholipase C is identified by the Enzyme Commission Number 3.1.4.11. The accepted name is phosphoinositide phospholipase C. -

Modeling Membrane-Protein Interactions
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 4 September 2018 doi:10.20944/preprints201809.0055.v1 Peer-reviewed version available at Biomolecules 2018, 8, 120; doi:10.3390/biom8040120 Review Modeling membrane-protein interactions Haleh Alimohamadi and Padmini Rangamani* Department of Mechanical and Aerospace Engineering, University of California San Diego, CA 92093, USA * Correspondence: [email protected]; Tel.: +1-858-534-4734 Abstract: In order to alter and adjust the shape of the membrane, cells harness various mechanisms of curvature generation. Many of these curvature generation mechanisms rely on the interactions between peripheral membrane 1 proteins, integral membrane proteins, and lipids in the bilayer membrane. One of the challenges in modeling these 2 processes is identifying the suitable constitutive relationships that describe the membrane free energy that includes 3 protein distribution and curvature generation capability. Here, we review some of the commonly used continuum elastic 4 membrane models that have been developed for this purpose and discuss their applications. Finally, we address some 5 fundamental challenges that future theoretical methods need to overcome in order to push the boundaries of current model 6 applications. 7 8 Keywords: Plasma membrane; Spontaneous curvature; Helfrich energy; Area difference elastic model; Protein crowding; Deviatoric curvature 9 10 11 1. Introduction 12 The ability of cellular membranes to bend and adapt their configurations is critical for a variety of cellular functions 13 including membrane trafficking processes [1,2], fission [3,4], fusion [5,6], differentiation [7], cell motility [8,9], and signal 14 transduction [10–12]. In order to dynamically reshape the membrane, cells rely on a variety of molecular mechanisms from 15 forces exerted by the cytoskeleton [13–15] and membrane-protein interactions [16–19]. -

ITPK1 Mediates the Lipid-Independent Synthesis of Inositol Phosphates Controlled by Metabolism
ITPK1 mediates the lipid-independent synthesis of inositol phosphates controlled by metabolism Yann Desfougèresa, Miranda S. C. Wilsona, Debabrata Lahaa, Gregory J. Millerb, and Adolfo Saiardia,1 aMedical Research Council Laboratory for Molecular Cell Biology, University College London, WC1E 6BT London, United Kingdom; and bDepartment of Chemistry, The Catholic University of America, Washington, DC 20064 Edited by Solomon H. Snyder, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved October 25, 2019 (received for review July 3, 2019) Inositol phosphates (IPs) comprise a network of phosphorylated The potential pathways to IP6-7-8 synthesis have a profound molecules that play multiple signaling roles in eukaryotes. IPs impact on how we interpret their signaling roles. If they are synthesis is believed to originate with IP3 generated from PIP2 by synthesized from IP3, their function has evolved in relation to phospholipase C (PLC). Here, we report that in mammalian cells lipid-dependent PLC and/or calcium signaling. This constraint is PLC-generated IPs are rapidly recycled to inositol, and uncover the relieved if IP6 synthesis occurs lipid-independently, originating enzymology behind an alternative “soluble” route to synthesis of directly from inositol, a “soluble” pathway. In yeast, which do not IPs. Inositol tetrakisphosphate 1-kinase 1 (ITPK1)—found in Asgard have IP3-regulated calcium signaling, IP6 synthesis strictly depends archaea, social amoeba, plants, and animals—phosphorylates on the PLC-generated IP3 (Figs. 1A and 2D). The higher com- I(3)P1 originating from glucose-6-phosphate, and I(1)P1 generated plexity of other eukaryotes might allow different pathways for IPs from sphingolipids, to enable synthesis of IP6. -

Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus Fumigatus
Journal of Fungi Article Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus Jizhou Li 1, Isabelle Mouyna 1, Christine Henry 1, Frédérique Moyrand 2, Christian Malosse 3, Julia Chamot-Rooke 3, Guilhem Janbon 2, Jean-Paul Latgé 1 and Thierry Fontaine 1,* 1 Unité des Aspergillus, 25 rue du Docteur Roux, Institut Pasteur, 25 rue du Docteur Roux, 75015 Paris, France; [email protected] (J.L.); [email protected] (I.M.); [email protected] (C.H.); [email protected] (J.-P.L.) 2 Unité de Biologie des ARN des Pathogènes Fongiques, Institut Pasteur, 25 rue du Docteur Roux, 75015 Paris, France; [email protected] (F.M.); [email protected] (G.J.) 3 Unité de Spectrométrie de Masse pour la Biologie, Institut Pasteur, CNRS USR 2000, 28 rue du Docteur Roux, 75015 Paris, France; [email protected] (C.M.); [email protected] (J.C.-R.) * Correspondence: [email protected]; Tel.: +33-145-688-358 Received: 8 December 2017; Accepted: 19 January 2018; Published: 2 Febuary 2018 Abstract: Glycosylphosphatidylinositols (GPIs) are lipid anchors allowing the exposure of proteins at the outer layer of the plasma membrane. In fungi, a number of GPI-anchored proteins (GPI-APs) are involved in the remodeling of the cell wall polymers. GPIs follow a specific biosynthetic pathway in the endoplasmic reticulum. After the transfer of the protein onto the GPI-anchor, a lipid remodeling occurs to substitute the diacylglycerol moiety by a ceramide. In addition to GPI-APs, A. fumigatus produces a GPI-anchored polysaccharide, the galactomannan (GM), that remains unique in the fungal kingdom. -

Dephosphorylation of 2,3-Bisphosphoglycerate by MIPP Expands the Regulatory Capacity of the Rapoport–Luebering Glycolytic Shunt
Dephosphorylation of 2,3-bisphosphoglycerate by MIPP expands the regulatory capacity of the Rapoport–Luebering glycolytic shunt Jaiesoon Cho*†, Jason S. King‡§, Xun Qian*, Adrian J. Harwood‡, and Stephen B. Shears*¶ *Laboratory of Signal Transduction, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Social Services, P.O. Box 12233, Research Triangle Park, NC 27709; and ‡Cardiff School of Biosciences, Cardiff University, Museum Avenue, Cardiff CF10 3US, United Kingdom Edited by Helen M. Ranney, University of California at San Diego, La Jolla, CA, and approved March 6, 2008 (received for review November 21, 2007) The Rapoport–Luebering glycolytic bypass comprises evolutionar- majority of mammals. By preferentially binding to deoxyhemo- ily conserved reactions that generate and dephosphorylate 2,3- globin, 2,3-BPG facilitates oxygen release from the erythrocyte bisphosphoglycerate (2,3-BPG). For >30 years, these reactions have to the surrounding tissues (6, 7). Thus, the regulation of eryth- been considered the responsibility of a single enzyme, the 2,3-BPG rocyte 2,3-BPG levels is key to efficiently meeting tissue oxygen synthase/2-phosphatase (BPGM). Here, we show that Dictyosteli- demands while also providing an important physiological adap- um, birds, and mammals contain an additional 2,3-BPG phospha- tation to oxygen deprivation (8), including that which occurs at tase that, unlike BPGM, removes the 3-phosphate. This discovery high altitude (9) or during postoperative anemia (10). reveals that the glycolytic pathway can bypass the formation of Despite the importance of carefully regulating 2,3-BPG turn- 3-phosphoglycerate, which is a precursor for serine biosynthesis over, little is known about how this might be achieved. -

Structural Basis for Phosphatidylinositol-Phosphate Biosynthesis
ARTICLE Received 8 Jun 2015 | Accepted 29 Aug 2015 | Published 16 Oct 2015 DOI: 10.1038/ncomms9505 OPEN Structural basis for phosphatidylinositol-phosphate biosynthesis Oliver B. Clarke1,*, David Tomasek2,*, Carla D. Jorge3,*, Meagan Belcher Dufrisne2, Minah Kim2, Surajit Banerjee4, Kanagalaghatta R. Rajashankar4, Lawrence Shapiro1, Wayne A. Hendrickson1, Helena Santos3 & Filippo Mancia2 Phosphatidylinositol is critical for intracellular signalling and anchoring of carbohydrates and proteins to outer cellular membranes. The defining step in phosphatidylinositol biosynthesis is catalysed by CDP-alcohol phosphotransferases, transmembrane enzymes that use CDP-diacylglycerol as donor substrate for this reaction, and either inositol in eukaryotes or inositol phosphate in prokaryotes as the acceptor alcohol. Here we report the structures of a related enzyme, the phosphatidylinositol-phosphate synthase from Renibacterium salmoninarum, with and without bound CDP-diacylglycerol to 3.6 and 2.5 Å resolution, respectively. These structures reveal the location of the acceptor site, and the molecular determinants of substrate specificity and catalysis. Functional characterization of the 40%-identical ortholog from Mycobacterium tuberculosis, a potential target for the development of novel anti-tuberculosis drugs, supports the proposed mechanism of substrate binding and catalysis. This work therefore provides a structural and functional framework to understand the mechanism of phosphatidylinositol-phosphate biosynthesis. 1 Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA. 2 Department of Physiology and Cellular Biophysics, Columbia University, New York, NY 10032, USA. 3 Biology Division, Instituto de Tecnologia Quı´mica e Biolo´gica, Universidade Nova de Lisboa, Avenida da Repu´blica-EAN, 2780-157 Oeiras, Portugal. 4 NE-CATand Department of Chemistry and Chemical Biology, Cornell University, Argonne National Laboratory, Argonne, IL 60439, USA. -

Forty Years Since the Structural Elucidation of Platelet-Activating Factor (PAF): Historical, Current, and Future Research Perspectives
molecules Review Forty Years Since the Structural Elucidation of Platelet-Activating Factor (PAF): Historical, Current, and Future Research Perspectives Ronan Lordan 1,2,* , Alexandros Tsoupras 1 , Ioannis Zabetakis 1,2 and Constantinos A. Demopoulos 3 1 Department of Biological Sciences, University of Limerick, V94 T9PX Limerick, Ireland; [email protected] (A.T.); [email protected] (I.Z.) 2 Health Research Institute (HRI), University of Limerick, V94 T9PX Limerick, Ireland 3 Department of Chemistry, National and Kapodistrian University of Athens, Panepistimioupolis, 15771 Athens, Greece; [email protected] * Correspondence: [email protected]; Tel.: +353-61-234-202 Academic Editor: Ferdinando Nicoletti Received: 2 November 2019; Accepted: 2 December 2019; Published: 3 December 2019 Abstract: In the late 1960s, Barbaro and Zvaifler described a substance that caused antigen induced histamine release from rabbit platelets producing antibodies in passive cutaneous anaphylaxis. Henson described a ‘soluble factor’ released from leukocytes that induced vasoactive amine release in platelets. Later observations by Siraganuan and Osler observed the existence of a diluted substance that had the capacity to cause platelet activation. In 1972, the term platelet-activating factor (PAF) was coined by Benveniste, Henson, and Cochrane. The structure of PAF was later elucidated by Demopoulos, Pinckard, and Hanahan in 1979. These studies introduced the research world to PAF, which is now recognised as a potent phospholipid mediator. Since its introduction to the literature, research on PAF has grown due to interest in its vital cell signalling functions and more sinisterly its role as a pro-inflammatory molecule in several chronic diseases including cardiovascular disease and cancer. -

Phosphatidylinositol 4,5-Bisphosphate Modifies Tubulin
The Journal of Neuroscience, March 1, 2002, 22(5):1668–1678 Phosphatidylinositol 4,5-Bisphosphate Modifies Tubulin  Participation in Phospholipase C 1 Signaling Juliana S. Popova,1 Arin K. Greene,1 Jia Wang,1 and Mark M. Rasenick1,2 Departments of 1Physiology and Biophysics and 2Psychiatry, University of Illinois at Chicago, College of Medicine, Chicago, Illinois 60612-7342 Tubulin forms the microtubule and regulates certain G-protein- the plasma membrane was demonstrated with confocal laser mediated signaling pathways. Both functions rely on the GTP- immunofluorescence microscopy. Although tubulin bound to ␣ ␣  binding properties of tubulin. Signal transduction through G q- both G q and PLC 1 , PIP2 facilitated the interaction between    ␣ regulated phospholipase C 1 (PLC 1 ) is activated by tubulin tubulin and PLC 1 but not that between tubulin and G q. ␣ ␣  through a direct transfer of GTP from tubulin to G q. However, However, PIP2 did augment formation of tubulin–G q–PLC 1   at high tubulin concentrations, inhibition of PLC 1 is observed. complexes. Subsequent to potentiating PLC 1 activation, sus-  This report demonstrates that tubulin inhibits PLC 1 by binding tained agonist-independent membrane binding of tubulin   ␣ the PLC 1 substrate phosphatidylinositol 4,5-bisphosphate at PIP2- and PLC 1-rich sites appeared to inhibit G q coupling  (PIP2 ). Tubulin binding of PIP2 was specific, because PIP2 but to PLC 1. Furthermore, colchicine increased membrane-  not phosphatidylinositol 3,4,5-trisphosphate, phosphatidylino- associated tubulin and also inhibited PLC 1 activity in SK- sitol 3-phosphate, phosphatidylinositol, phosphatidylcholine, N-SH cells. Thus, tubulin, depending on local membrane con- phosphatidylethanolamine, or inositol 1,4,5-trisphosphate in- centration, may serve as a positive or negative regulator of hibited microtubule assembly.