Temperature Modulation of Biological Clocks in a Reef-Building Coral
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Domain Stationid Station UDC Performance Date
Number of days Amount Amount Performance Total Per Domain StationId Station UDC processed for from from Public Date Minute Rate distribution Broadcast Reception RADIO BR ONE BBC RADIO 1 NON PEAK BRA01 CENSUS 92 7.8347 4.2881 3.5466 RADIO BR ONE BBC RADIO 1 LOW PEAK BRB01 CENSUS 92 10.7078 7.1612 3.5466 RADIO BR ONE BBC RADIO 1 HIGH PEAK BRC01 CENSUS 92 13.5380 9.9913 3.5466 RADIO BR TWO BBC RADIO 2 NON PEAK BRA02 CENSUS 92 17.4596 11.2373 6.2223 RADIO BR TWO BBC RADIO 2 LOW PEAK BRB02 CENSUS 92 24.9887 18.7663 6.2223 RADIO BR TWO BBC RADIO 2 HIGH PEAK BRC02 CENSUS 92 32.4053 26.1830 6.2223 RADIO BR1EXT BBC RADIO 1XTRA NON PEAK BRA10 CENSUS 92 1.4814 1.4075 0.0739 RADIO BR1EXT BBC RADIO 1XTRA LOW PEAK BRB10 CENSUS 92 2.4245 2.3506 0.0739 RADIO BR1EXT BBC RADIO 1XTRA HIGH PEAK BRC10 CENSUS 92 3.3534 3.2795 0.0739 RADIO BRASIA BBC ASIAN NETWORK NON PEAK BRA65 CENSUS 92 1.4691 1.4593 0.0098 RADIO BRASIA BBC ASIAN NETWORK LOW PEAK BRB65 CENSUS 92 2.4468 2.4371 0.0098 RADIO BRASIA BBC ASIAN NETWORK HIGH PEAK BRC65 CENSUS 92 3.4100 3.4003 0.0098 RADIO BRBEDS BBC THREE COUNTIES RADIO NON PEAK BRA62 CENSUS 92 0.1516 0.1104 0.0411 RADIO BRBEDS BBC THREE COUNTIES RADIO LOW PEAK BRB62 CENSUS 92 0.2256 0.1844 0.0411 RADIO BRBEDS BBC THREE COUNTIES RADIO HIGH PEAK BRC62 CENSUS 92 0.2985 0.2573 0.0411 RADIO BRBERK BBC RADIO BERKSHIRE NON PEAK BRA64 CENSUS 92 0.0803 0.0569 0.0233 RADIO BRBERK BBC RADIO BERKSHIRE LOW PEAK BRB64 CENSUS 92 0.1184 0.0951 0.0233 RADIO BRBERK BBC RADIO BERKSHIRE HIGH PEAK BRC64 CENSUS 92 0.1560 0.1327 0.0233 RADIO BRBRIS BBC -

Stations Monitored
Stations Monitored 10/01/2019 Format Call Letters Market Station Name Adult Contemporary WHBC-FM AKRON, OH MIX 94.1 Adult Contemporary WKDD-FM AKRON, OH 98.1 WKDD Adult Contemporary WRVE-FM ALBANY-SCHENECTADY-TROY, NY 99.5 THE RIVER Adult Contemporary WYJB-FM ALBANY-SCHENECTADY-TROY, NY B95.5 Adult Contemporary KDRF-FM ALBUQUERQUE, NM 103.3 eD FM Adult Contemporary KMGA-FM ALBUQUERQUE, NM 99.5 MAGIC FM Adult Contemporary KPEK-FM ALBUQUERQUE, NM 100.3 THE PEAK Adult Contemporary WLEV-FM ALLENTOWN-BETHLEHEM, PA 100.7 WLEV Adult Contemporary KMVN-FM ANCHORAGE, AK MOViN 105.7 Adult Contemporary KMXS-FM ANCHORAGE, AK MIX 103.1 Adult Contemporary WOXL-FS ASHEVILLE, NC MIX 96.5 Adult Contemporary WSB-FM ATLANTA, GA B98.5 Adult Contemporary WSTR-FM ATLANTA, GA STAR 94.1 Adult Contemporary WFPG-FM ATLANTIC CITY-CAPE MAY, NJ LITE ROCK 96.9 Adult Contemporary WSJO-FM ATLANTIC CITY-CAPE MAY, NJ SOJO 104.9 Adult Contemporary KAMX-FM AUSTIN, TX MIX 94.7 Adult Contemporary KBPA-FM AUSTIN, TX 103.5 BOB FM Adult Contemporary KKMJ-FM AUSTIN, TX MAJIC 95.5 Adult Contemporary WLIF-FM BALTIMORE, MD TODAY'S 101.9 Adult Contemporary WQSR-FM BALTIMORE, MD 102.7 JACK FM Adult Contemporary WWMX-FM BALTIMORE, MD MIX 106.5 Adult Contemporary KRVE-FM BATON ROUGE, LA 96.1 THE RIVER Adult Contemporary WMJY-FS BILOXI-GULFPORT-PASCAGOULA, MS MAGIC 93.7 Adult Contemporary WMJJ-FM BIRMINGHAM, AL MAGIC 96 Adult Contemporary KCIX-FM BOISE, ID MIX 106 Adult Contemporary KXLT-FM BOISE, ID LITE 107.9 Adult Contemporary WMJX-FM BOSTON, MA MAGIC 106.7 Adult Contemporary WWBX-FM -

Latin America Spanish Only
Newswire.com LLC 5 Penn Plaza, 23rd Floor| New York, NY 10001 Telephone: 1 (800) 713-7278 | www.newswire.com Latin America Spanish Only Distribution to online destinations, including media and industry websites and databases, through proprietary and news agency networks (DyN and Notimex). In addition, the circuit features the following complimentary added-value services: • Posting to online services and portals. • Coverage on Newswire's media-only website and custom push email service, Newswire for Journalists, reaching 100,000 registered journalists from more than 170 countries and in more than 40 different languages. • Distribution of listed company news to financial professionals around the world via Thomson Reuters, Bloomberg and proprietary networks. Comprehensive newswire distribution to news media in 19 Central and South American countries: Argentina, Bolivia, Chile, Colombia, Costa Rica, Cuba, Domincan Republic, Ecuador, El Salvador, Guatemala, Honduras, Mexico, Nicaragua, Panama, Paraguay, Peru, Puerto Rico, Uruguay and Venezuela. Translated and distributed in Spanish. Please note that this list is intended for general information purposes and may adjust from time to time without notice. 4,028 Points Country Media Point Media Type Argentina 0223.com.ar Online Argentina Acopiadores de Córdoba Online Argentina Agensur.info (Agencia de Noticias del Mercosur) Agencies Argentina AgriTotal.com Online Argentina Alfil Newspaper Argentina Amdia blog Blog Argentina ANRed (Agencia de Noticias Redacción) Agencies Argentina Argentina Ambiental -

Preuzimanje Preuzimanje PDF-A
VAŠA UPUTSTVA ZA UPOTREBU Digitalno uputstvo u vozilu Proučite sadržaje uputstva za upotrebu direktno u multimedijalnom sistemu vašeg vozila (tačka menija „Vozilo“). Počnite od kratkog uputstva ili produbite svoje znanje korisnim savetima. Torbica za papire vozila Ovde možete da pronađete informacije o aktiviranju, servisna uputstva i garanciju za vaše vozilo u štampanom obliku. Digitalno uputstvo na internetu Uputstvo za upotrebu možete da pronađete na Mercedes-Benz veb-stranici. Digitalno uputstvo kao aplikacija Aplikacija Mercedes-Benz Guides je dostupna u najpoznatijim internet prodavni‐ cama aplikacija. Sprinter Uputstvo za upotrebu É9075844602Z119@ËÍ 9075844602Z119 Broj porudžbine T907 0082 19 Broj dela 907 584 46 02 Z119 Izdanje 2018-03a m Mercedes-Benz SprinterUputstvo za upotrebu Simboli Impresum U ovom uputstvu za rukovanje možete pronaći Internet sledeće simbole: Dodatne informacije o Mercedes-Benz vozilima i kompaniji Daimler AG možete da pronađete na & OPASNOST Opasnost zbog nepoštovanja internetu, na adresi: bezbednosnih napomena http://www.mercedes-benz.com Upozoravajuće napomene skreću pažnju na http://www.daimler.com opasnosti koje mogu da ugroze vaše zdravlje ili život, odnosno zdravlje ili život drugih Redakcija osoba. Pitanja i sugestije u vezi sa ovim uputstvom za upotrebu možete da pošaljete redakciji za teh‐ # Poštujte bezbednosne napomene u ničku dokumentaciju na sledeću adresu: ovom uputstvu. Daimler AG, HPC: CAC, Customer Service, 70546 Stuttgart, Deutschland + NAPOMENA ZA ZAŠTITU ŽIVOTNE SRE‐ DINE Zagađenje okoline usled nepoštova‐ ©Daimler AG: Doštampavanje, prevođenje i nja napomena za zaštitu životne sredine. umnožavanje, čak i samo delova ovog uputstva, nisu dozvoljeni bez pisane dozvole kompanije Dai‐ Napomene za zaštitu životne sredine pružaju mler AG. informacije o postupanju prema ekološkim zahtevima ili ekološkom odlaganju otpada. -

Evaluating MTV Emas 2011 with Belfast Music Week
Evaluating MTV EMAs 2011 with Belfast Music Week EVALUATING MTV EMAS 2011 WITH BELFAST MUSIC WEEK 1 The week the world came to Belfast Belfast Music Week featuring the MTV EMAs 2011 The 2011 MTV EMAs, alongside Belfast Music Week, was a landmark event for Belfast - placing the city firmly on the world musical stage. And as is only fitting for a world class event, the figures are pretty impressive: • 1.2 billion – the number of people who watched the MTV EMAs across the globe • 669 million – the number of media news opportunities generated • £22 million – the total economic impact for Belfast • £10 million – the amount of additional tourism revenue for the city • 8,000 – the number of room nights booked in Belfast hotels • 2,399 – the number of media articles released • 100% - the percentage of people who would recommend attending Belfast Music Week events • 33,500 - the number of people who attended Belfast Music Week • 170 - the number of gigs during Belfast Music Week The EMAs brought the biggest international acts to Belfast. And it wasn’t just the people of Belfast who were inspired by The ‘who’s who’ of the pop world, including Coldplay, Lady the EMAs. The world’s media arrived at our door expecting Gaga, Bruno Mars, Queen, Jessie J and Justin Bieber all the professionalism associated with such a high profile appeared at the city’s Odyssey Arena. event. Once again we did not disappoint. The enthusiasm of Belfast’s people was felt around the globe through press Meanwhile, local heroes Snow Patrol played a free open-air articles, webcasts and television broadcasts. -

Basisdaten 2016 1
Verzeichnis der Tabellen und Grafiken Media Perspektiven Basisdaten 2016 1 Seite Rundfunk: Programmangebot und Empfangssituation TV-Haushalte nach Empfangsebenen in Deutschland 2016 4 Empfangspotenzial der deutschen Fernsehsender 2016 4 Öffentlich-rechtlicher Rundfunk: Erträge/Leistungen Rundfunkgebühren/Rundfunkbeitrag 6 Erträge aus der Rundfunkgebühr bzw. dem Rundfunkbeitrag 7 Werbefunkumsätze der ARD-Werbung 7 Werbefernsehumsätze von ARD und ZDF 7 Programmleistung der ARD 2015: Erstes Fernsehprogramm 8 Programmleistung von ARD und ZDF für KiKA und Phoenix 2015 8 Programmleistung von ARD und ZDF für Arte 2015 9 Programmleistung des ZDF 2015 9 Programmleistung von 3sat 2015 10 Programmleistung von Deutschlandradio 2015 10 Programmleistung der Deutschen Welle 2015 11 Programmleistung der ARD 2015: Hörfunk 11 Privater Rundfunk: Erträge/Leistungen Werbeumsätze privater Hörfunkanbieter 12 Bruttowerbeumsätze privater Fernsehanbieter 12 Programmleistung von RTL 2015 13 Programmleistung von ProSieben 2015 14 Programmleistung von Sat.1 2015 14 Programmleistung von VOX 2015 14 Programmleistung von Super RTL 2015 15 Programmleistung von RTL II 2015 15 Programmleistung von kabel eins 2015 16 Programmleistung von Sport1 2015 16 Programmprofile im dualen Rundfunksystem Spartenprofile von Das Erste, ZDF, RTL, Sat.1 und ProSieben 2013 bis 2015 17 Programmstruktur 2015: Sparten und Formen von Das Erste, ZDF, RTL, Sat.1 und ProSieben 19 Themenstruktur der wichtigsten Nachrichtensendungen von ARD, ZDF, RTL und Sat.1 22 Themenkategorien und ausgewählte -

2017 Civil Society Organization Sustainability Index
STRENGTHENING STRENGTHENING CIVIL SOCIETY CIVIL SOCIETY GLOBALLY GLOBALLY 2017 CIVIL SOCIETY ORGANIZATION SUSTAINABILITY INDEX FOR SUB-SAHARAN AFRICA 9th EDITION - DECEMBER 2018 2017 CIVIL SOCIETY ORGANIZATION SUSTAINABILITY INDEX FOR SUB-SAHARAN AFRICA 9th EDITION - DECEMBER 2018 Developed By: United States Agency for International Development Bureau for Democracy, Conflict and Humanitarian Assistance Center of Excellence on Democracy, Human Rights and Governance In Partnership With: FHI 360 International Center for Not-for-Profit Law (ICNL) Acknowledgment: This publication was made possible through support provided by the United States Agency for International Development (USAID) under Cooperative Agreement No. AID-OAA-LA-17-00003. Disclaimer: The opinions expressed herein are those of the panelists and other project researchers and do not necessarily reflect the views of USAID or FHI 360. Cover Photo: Fanis Lisiagali, Executive Director of Healthcare Assistance Kenya, leads the White Ribbon Campaign in a march in Nairobi to promote its rapid response call center hotline, which responds to violence against women in elections (October 2017). Photo Credit: Carla Chianese, Kenya Electoral Assistance Program, International Foundation for Electoral Systems TABLE OF CONTENTS INTRODUCTION .................................................................................................................................................... i ACKNOWLEDGMENTS ....................................................................................................................................... -

QUARTERLY SUMMARY of RADIO LISTENING Survey Period Ending 15Th September 2019
QUARTERLY SUMMARY OF RADIO LISTENING Survey Period Ending 15th September 2019 PART 1 - UNITED KINGDOM (INCLUDING CHANNEL ISLANDS AND ISLE OF MAN) Adults aged 15 and over: population 55,032,000 Survey Weekly Reach Average Hours Total Hours Share in Period '000 % per head per listener '000 TSA % All Radio Q 48537 88 18.0 20.4 989221 100.0 All BBC Radio Q 33451 61 8.9 14.6 488274 49.4 All BBC Radio 15-44 Q 12966 51 4.6 8.9 115944 33.9 All BBC Radio 45+ Q 20485 69 12.5 18.2 372330 57.5 All BBC Network Radio1 Q 30828 56 7.7 13.8 425563 43.0 BBC Local Radio Q 7430 14 1.1 8.4 62711 6.3 All Commercial Radio Q 35930 65 8.6 13.2 475371 48.1 All Commercial Radio 15-44 Q 17884 71 8.5 12.0 214585 62.7 All Commercial Radio 45+ Q 18046 61 8.8 14.5 260786 40.3 All National Commercial1 Q 22361 41 3.8 9.5 211324 21.4 All Local Commercial (National TSA) Q 25988 47 4.8 10.2 264047 26.7 Other Radio Q 4035 7 0.5 6.3 25577 2.6 Source: RAJAR/Ipsos MORI/RSMB 1 See note on back cover. For survey periods and other definitions please see back cover. Please note that the information contained within this quarterly data release has yet to be announced or otherwise made public Embargoed until 00.01 am and as such could constitute relevant information for the purposes of section 118 of FSMA and non-public price sensitive 24th October 2019 information for the purposes of the Criminal Justice Act 1993. -

Media Perspektiven Basisdaten
ISSN 0942-072X Media Perspektiven Basisdaten Daten zur Mediensituation in Deutschland 2020 Rundfunk: Programmangebot und Empfangssituation Öffentlich-rechtlicher Rundfunk: Erträge/Leistungen Privater Rundfunk: Erträge/Leistungen Programmprofile im dualen Rundfunksystem Medienkonzerne: Beteiligungen Presse, Buch Kino/Film und Video/DVD Theater Unterhaltungselektronik, Musikmedien Mediennutzung Werbung Allgemeine Daten Media Perspektiven Basisdaten 2020 Media Perspektiven Basisdaten Daten zur Mediensituation in Deutschland 2020 In dieser jährlich aktualisierten Publikation werden Basisdaten zum gesamten Mediensektor zusammengestellt. Berücksichtigt werden Hörfunk und Fernsehen, Presse, der Buchmarkt, Kino/Film, Video/DVD, Theater, Unterhaltungselektronik/Musikmedien sowie Werbung. Weitere Schwerpunkte sind die Beteiligungen und Verflechtungen der großen Medienkonzerne sowie die Nutzung der tagesaktuellen Medien Fernsehen, Radio, Presse und Internet. In der Sammlung werden nur kontinuierlich erhobene Datenquellen berücksichtigt, um Entwicklungen im Zeitverlauf dokumentieren zu können. Frankfurt am Main, Februar 2021 Datenrecherche: Michael Braband Redaktion: Hanna Puffer Verzeichnis der Tabellen und Grafiken Media Perspektiven Basisdaten 2020 1 Seite Rundfunk: Programmangebot und Empfangssituation TV-Haushalte nach Empfangsebenen in Deutschland 2020 4 Empfangspotenzial der deutschen Fernsehsender 2020 4 Öffentlich-rechtlicher Rundfunk: Erträge/Leistungen Rundfunkgebühren/Rundfunkbeitrag 6 Erträge aus der Rundfunkgebühr bzw. dem Rundfunkbeitrag -
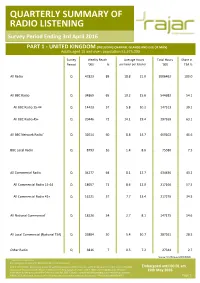
QUARTERLY SUMMARY of RADIO LISTENING Survey Period Ending 3Rd April 2016
QUARTERLY SUMMARY OF RADIO LISTENING Survey Period Ending 3rd April 2016 PART 1 - UNITED KINGDOM (INCLUDING CHANNEL ISLANDS AND ISLE OF MAN) Adults aged 15 and over: population 53,575,000 Survey Weekly Reach Average Hours Total Hours Share in Period '000 % per head per listener '000 TSA % All Radio Q 47823 89 18.8 21.0 1006462 100.0 All BBC Radio Q 34869 65 10.2 15.6 544682 54.1 All BBC Radio 15-44 Q 14423 57 5.8 10.2 147513 39.1 All BBC Radio 45+ Q 20446 72 14.1 19.4 397169 63.1 All BBC Network Radio1 Q 32014 60 8.8 14.7 469102 46.6 BBC Local Radio Q 8793 16 1.4 8.6 75580 7.5 All Commercial Radio Q 34277 64 8.1 12.7 434436 43.2 All Commercial Radio 15-44 Q 18057 71 8.6 12.0 217166 57.5 All Commercial Radio 45+ Q 16221 57 7.7 13.4 217270 34.5 All National Commercial1 Q 18220 34 2.7 8.1 147175 14.6 All Local Commercial (National TSA) Q 26884 50 5.4 10.7 287261 28.5 Other Radio Q 3816 7 0.5 7.2 27344 2.7 Source: RAJAR/Ipsos MORI/RSMB 1 See note on back cover. For survey periods and other definitions please see back cover. Please note that the information contained within this quarterly data release has yet to be announced or otherwise made public Embargoed until 00.01 am and as such could constitute relevant information for the purposes of section 118 of FSMA and non-public price sensitive 19th May 2016 information for the purposes of the Criminal Justice Act 1993. -

PRS Radio Dec 2018.Xlsx
No of Days in Total Per Amount from Amount from Domain StationId Station UDC Performance Date Period Minute Rate Broadcast Public Reception RADIO BR ONE BBC RADIO 1 B0001 31/12/2099 92 £12.2471 £7.3036 £4.9435 RADIO BR TWO BBC RADIO 2 B0002 31/12/2099 92 £25.4860 £25.3998 £0.0862 RADIO BR1EXT BBC 1XTRA CENSUS B0106 31/12/2099 92 £2.8113 £2.7199 £0.0914 RADIO BRASIA BBC ASIAN NETWORK (CENSUS) B0064 31/12/2099 92 £3.6951 £3.3058 £0.3892 RADIO BRBEDS BBC THREE COUNTIES RADIO (CENSUS) B0065 31/12/2099 92 £0.2445 £0.2442 £0.0003 RADIO BRBERK BBC RADIO BERKSHIRE (CENSUS) B0103 31/12/2099 92 £0.1436 £0.1435 £0.0002 RADIO BRBRIS BBC RADIO BRISTOL (CENSUS) B0066 31/12/2099 92 £0.1532 £0.1531 £0.0002 RADIO BRCAMB BBC RADIO CAMBRIDGESHIRE (CENSUS) B0067 31/12/2099 92 £0.1494 £0.1493 £0.0002 RADIO BRCLEV BBC RADIO TEES (CENSUS) B0068 31/12/2099 92 £0.1478 £0.1477 £0.0002 RADIO BRCMRU BBC RADIO CYMRU B0011 31/12/2099 92 £0.5707 £0.5690 £0.0017 RADIO BRCORN BBC RADIO CORNWALL (CENSUS) B0069 31/12/2099 92 £0.1535 £0.1534 £0.0002 RADIO BRCOVN BBC RADIO COVENTRY AND WATWICKSHIRE(CENSUS) B0070 31/12/2099 92 £0.1023 £0.1022 £0.0001 RADIO BRCUMB BBC RADIO CUMBRIA (CENSUS) B0071 31/12/2099 92 £0.1085 £0.1084 £0.0001 RADIO BRCYMM BBC RADIO CYMRU 2 B0114 31/12/2099 92 £0.5707 £0.5690 £0.0017 RADIO BRDEVN BBC RADIO DEVON (CENSUS) B0072 31/12/2099 92 £0.2421 £0.2419 £0.0003 RADIO BRDRBY BBC RADIO DERBY (CENSUS) B0073 31/12/2099 92 £0.1535 £0.1534 £0.0002 RADIO BRESSX BBC ESSEX (CENSUS) B0074 31/12/2099 92 £0.2091 £0.2089 £0.0002 RADIO BRFIVE BBC FIVE LIVE B0005 -

2017 Civil Society Organization Sustainability Index
STRENGTHENING STRENGTHENING CIVIL SOCIETY CIVIL SOCIETY GLOBALLY GLOBALLY 2017 CIVIL SOCIETY ORGANIZATION SUSTAINABILITY INDEX FOR SUB-SAHARAN AFRICA 9th EDITION - DECEMBER 2018 2017 CIVIL SOCIETY ORGANIZATION SUSTAINABILITY INDEX FOR SUB-SAHARAN AFRICA 9th EDITION - DECEMBER 2018 Developed By: United States Agency for International Development Bureau for Democracy, Conflict and Humanitarian Assistance Center of Excellence on Democracy, Human Rights and Governance In Partnership With: FHI 360 International Center for Not-for-Profit Law (ICNL) Acknowledgment: This publication was made possible through support provided by the United States Agency for International Development (USAID) under Cooperative Agreement No. AID-OAA-LA-17-00003. Disclaimer: The opinions expressed herein are those of the panelists and other project researchers and do not necessarily reflect the views of USAID or FHI 360. Cover Photo: Fanis Lisiagali, Executive Director of Healthcare Assistance Kenya, leads the White Ribbon Campaign in a march in Nairobi to promote its rapid response call center hotline, which responds to violence against women in elections (October 2017). Photo Credit: Carla Chianese, Kenya Electoral Assistance Program, International Foundation for Electoral Systems TABLE OF CONTENTS INTRODUCTION .................................................................................................................................................... i ACKNOWLEDGMENTS .......................................................................................................................................