Evaluating Insect-Host Interactions As a Driver of Species Divergence in Palm Flower Weevils
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Journal of the International Palm Society Vol. 58(4) Dec. 2014 the INTERNATIONAL PALM SOCIETY, INC
Palms Journal of the International Palm Society Vol. 58(4) Dec. 2014 THE INTERNATIONAL PALM SOCIETY, INC. The International Palm Society Palms (formerly PRINCIPES) Journal of The International Palm Society Founder: Dent Smith The International Palm Society is a nonprofit corporation An illustrated, peer-reviewed quarterly devoted to engaged in the study of palms. The society is inter- information about palms and published in March, national in scope with worldwide membership, and the June, September and December by The International formation of regional or local chapters affiliated with the Palm Society Inc., 9300 Sandstone St., Austin, TX international society is encouraged. Please address all 78737-1135 USA. inquiries regarding membership or information about Editors: John Dransfield, Herbarium, Royal Botanic the society to The International Palm Society Inc., 9300 Gardens, Kew, Richmond, Surrey, TW9 3AE, United Sandstone St., Austin, TX 78737-1135 USA, or by e-mail Kingdom, e-mail [email protected], tel. 44-20- to [email protected], fax 512-607-6468. 8332-5225, Fax 44-20-8332-5278. OFFICERS: Scott Zona, Dept. of Biological Sciences (OE 167), Florida International University, 11200 SW 8 Street, President: Leland Lai, 21480 Colina Drive, Topanga, Miami, Florida 33199 USA, e-mail [email protected], tel. California 90290 USA, e-mail [email protected], 1-305-348-1247, Fax 1-305-348-1986. tel. 1-310-383-2607. Associate Editor: Natalie Uhl, 228 Plant Science, Vice-Presidents: Jeff Brusseau, 1030 Heather Drive, Cornell University, Ithaca, New York 14853 USA, e- Vista, California 92084 USA, e-mail mail [email protected], tel. 1-607-257-0885. -

Coleoptera) (Excluding Anthribidae
A FAUNAL SURVEY AND ZOOGEOGRAPHIC ANALYSIS OF THE CURCULIONOIDEA (COLEOPTERA) (EXCLUDING ANTHRIBIDAE, PLATPODINAE. AND SCOLYTINAE) OF THE LOWER RIO GRANDE VALLEY OF TEXAS A Thesis TAMI ANNE CARLOW Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 1997 Major Subject; Entomology A FAUNAL SURVEY AND ZOOGEOGRAPHIC ANALYSIS OF THE CURCVLIONOIDEA (COLEOPTERA) (EXCLUDING ANTHRIBIDAE, PLATYPODINAE. AND SCOLYTINAE) OF THE LOWER RIO GRANDE VALLEY OF TEXAS A Thesis by TAMI ANNE CARLOW Submitted to Texas AgcM University in partial fulltllment of the requirements for the degree of MASTER OF SCIENCE Approved as to style and content by: Horace R. Burke (Chair of Committee) James B. Woolley ay, Frisbie (Member) (Head of Department) Gilbert L. Schroeter (Member) August 1997 Major Subject: Entomology A Faunal Survey and Zoogeographic Analysis of the Curculionoidea (Coleoptera) (Excluding Anthribidae, Platypodinae, and Scolytinae) of the Lower Rio Grande Valley of Texas. (August 1997) Tami Anne Carlow. B.S. , Cornell University Chair of Advisory Committee: Dr. Horace R. Burke An annotated list of the Curculionoidea (Coleoptem) (excluding Anthribidae, Platypodinae, and Scolytinae) is presented for the Lower Rio Grande Valley (LRGV) of Texas. The list includes species that occur in Cameron, Hidalgo, Starr, and Wigacy counties. Each of the 23S species in 97 genera is tteated according to its geographical range. Lower Rio Grande distribution, seasonal activity, plant associations, and biology. The taxonomic atTangement follows O' Brien &, Wibmer (I og2). A table of the species occuning in patxicular areas of the Lower Rio Grande Valley, such as the Boca Chica Beach area, the Sabal Palm Grove Sanctuary, Bentsen-Rio Grande State Park, and the Falcon Dam area is included. -

The Beetle Fauna of Dominica, Lesser Antilles (Insecta: Coleoptera): Diversity and Distribution
INSECTA MUNDI, Vol. 20, No. 3-4, September-December, 2006 165 The beetle fauna of Dominica, Lesser Antilles (Insecta: Coleoptera): Diversity and distribution Stewart B. Peck Department of Biology, Carleton University, 1125 Colonel By Drive, Ottawa, Ontario K1S 5B6, Canada stewart_peck@carleton. ca Abstract. The beetle fauna of the island of Dominica is summarized. It is presently known to contain 269 genera, and 361 species (in 42 families), of which 347 are named at a species level. Of these, 62 species are endemic to the island. The other naturally occurring species number 262, and another 23 species are of such wide distribution that they have probably been accidentally introduced and distributed, at least in part, by human activities. Undoubtedly, the actual numbers of species on Dominica are many times higher than now reported. This highlights the poor level of knowledge of the beetles of Dominica and the Lesser Antilles in general. Of the species known to occur elsewhere, the largest numbers are shared with neighboring Guadeloupe (201), and then with South America (126), Puerto Rico (113), Cuba (107), and Mexico-Central America (108). The Antillean island chain probably represents the main avenue of natural overwater dispersal via intermediate stepping-stone islands. The distributional patterns of the species shared with Dominica and elsewhere in the Caribbean suggest stages in a dynamic taxon cycle of species origin, range expansion, distribution contraction, and re-speciation. Introduction windward (eastern) side (with an average of 250 mm of rain annually). Rainfall is heavy and varies season- The islands of the West Indies are increasingly ally, with the dry season from mid-January to mid- recognized as a hotspot for species biodiversity June and the rainy season from mid-June to mid- (Myers et al. -
Winter-Fall Sale 2002 Palm Trees-Web
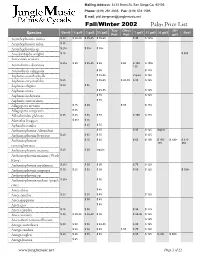
Mailing Address: 3233 Brant St. San Diego Ca, 92103 Phone: (619) 291 4605 Fax: (619) 574 1595 E mail: [email protected] Fall/Winter 2002 Palm Price List Tree Citrus 25/+ Band$ 1 gal$ 2 gal$ 3/5 gal$ 7 gal$ 15 gal$ 20 gal$ Box$ Species Pot$ Pot$ gal$ Acanthophoenix crinita $ 30 $ 30-40 $ 35-45 $ 55-65 $ 95 $ 125+ Acanthophoenix rubra $ 35 Acanthophoenix sp. $ 25+ $ 35+ $ 55+ Acoelorrhaphe wrightii $ 15 $ 300 Acrocomia aculeata $ 25+ $ 35 $ 35-45 $ 65 $ 65 $ 100- $ 150+ Actinokentia divaricata 135 Actinorhytis calapparia $ 55 $ 125 Aiphanes acanthophylla $ 45-55 inquire $ 125 Aiphanes caryotaefolia $ 25 $ 55-65 $ 45-55 $ 85 $ 125 Aiphanes elegans $ 20 $ 35 Aiphanes erosa $ 45-55 $ 125 Aiphanes lindeniana $ 55 $ 125 Aiphanes vincentsiana $ 55 Allagoptera arenaria $ 25 $ 40 $ 55 $ 135 Allagoptera campestris $ 35 Alloschmidtia glabrata $ 35 $ 45 $ 55 $ 85 $ 150 $ 175 Alsmithia longipes $ 35+ $ 55 Aphandra natalia $ 35 $ 55 Archontophoenix Alexandrae $ 55 $ 85 $ 125 inquire Archontophoenix Beatricae $ 20 $ 35 $ 55 $ 125 Archontophoenix $ 25 $ 45 $ 65 $ 100 $ 150- $ 200+ $ 310- 175 350 cunninghamiana Archontophoenix maxima $ 25 $ 30 inquire Archontophoenix maxima (Wash River) Archontophoenix myolaensis $ 25+ $ 30 $ 50 $ 75 $ 125 Archontophoenix purpurea $ 30 $ 25 $ 35 $ 50 $ 85 $ 125 $ 300+ Archontophoenix sp. Archontophoenix tuckerii (peach $ 25+ $ 55 river) Areca alicae $ 45 Areca catechu $ 20 $ 35 $ 45 $ 125 Areca guppyana $ 30 $ 45 Areca ipot $ 45 Areca triandra $ 25 $ 30 $ 95 $ 125 Areca vestiaria $ 25 $ 30-35 $ 35-40 $ 55 $ 85-95 $ 125 Arecastrum romanzoffianum $ 125 Arenga australasica $ 20 $ 30 $ 35 $ 45-55 $ 85 $ 125 Arenga caudata $ 20 $ 30 $ 45 $ 55 $ 75 $ 100 Arenga engleri $ 20 $ 60 $ 35 $ 45 $ 85 $ 125 $ 200 $ 300+ Arenga hastata $ 25 www.junglemusic.net Page 1 of 22 Tree Citrus 25/+ Band$ 1 gal$ 2 gal$ 3/5 gal$ 7 gal$ 15 gal$ 20 gal$ Box$ Species Pot$ Pot$ gal$ Arenga hookeriana inquire Arenga micranthe 'Lhutan' $ 20 inquire Arenga pinnata $ 35 $ 50 $ 85 $ 125 Arenga sp. -

The Curculionoidea of the Maltese Islands (Central Mediterranean) (Coleoptera)
BULLETIN OF THE ENTOMOLOGICAL SOCIETY OF MALTA (2010) Vol. 3 : 55-143 The Curculionoidea of the Maltese Islands (Central Mediterranean) (Coleoptera) David MIFSUD1 & Enzo COLONNELLI2 ABSTRACT. The Curculionoidea of the families Anthribidae, Rhynchitidae, Apionidae, Nanophyidae, Brachyceridae, Curculionidae, Erirhinidae, Raymondionymidae, Dryophthoridae and Scolytidae from the Maltese islands are reviewed. A total of 182 species are included, of which the following 51 species represent new records for this archipelago: Araecerus fasciculatus and Noxius curtirostris in Anthribidae; Protapion interjectum and Taeniapion rufulum in Apionidae; Corimalia centromaculata and C. tamarisci in Nanophyidae; Amaurorhinus bewickianus, A. sp. nr. paganettii, Brachypera fallax, B. lunata, B. zoilus, Ceutorhynchus leprieuri, Charagmus gressorius, Coniatus tamarisci, Coniocleonus pseudobliquus, Conorhynchus brevirostris, Cosmobaris alboseriata, C. scolopacea, Derelomus chamaeropis, Echinodera sp. nr. variegata, Hypera sp. nr. tenuirostris, Hypurus bertrandi, Larinus scolymi, Leptolepurus meridionalis, Limobius mixtus, Lixus brevirostris, L. punctiventris, L. vilis, Naupactus cervinus, Otiorhynchus armatus, O. liguricus, Rhamphus oxyacanthae, Rhinusa antirrhini, R. herbarum, R. moroderi, Sharpia rubida, Sibinia femoralis, Smicronyx albosquamosus, S. brevicornis, S. rufipennis, Stenocarus ruficornis, Styphloderes exsculptus, Trichosirocalus centrimacula, Tychius argentatus, T. bicolor, T. pauperculus and T. pusillus in Curculionidae; Sitophilus zeamais and -

Essential Oils and Oil from Seeds of Syagrus Coronata
Vol. 10(23), pp. 310-317, 17 June, 2016 DOI: 10.5897/JMPR2016.6098 Article Number: 89CD55858997 ISSN 1996-0875 Journal of Medicinal Plants Research Copyright © 2016 Author(s) retain the copyright of this article http://www.academicjournals.org/JMPR Full Length Research Paper Syagrus coronata seed oils have antimicrobial action against multidrug-resistant Staphylococcus aureus Cibele Maria Alves da Silva Bessa1, Rodrigo Santana do Nascimento1, Renata Carla Corrêa Alves1*, José Matias Anselmo2, Ana Paula Sant'Anna da Silva1, Alexandre Gomes da Silva1, Vera Lúcia de Menezes Lima1, Josean Fechine Tavares3, Luís Cláudio Nascimento da Silva1,2, Márcia Vanusa da Silva1 and Maria Tereza dos Santos Correia1 1Departamento de Bioquímica, Centro de Ciências Biológicas, Universidade Federal de Pernambuco, Av. Professor Moraes Rego, Cidade Universitária, 1235, 50670-901, Recife, Pernambuco, Brazil. 2Faculdade Pernambucana de Saúde, Av. Jean Emile Favre, 420, Imbiribeira, 51200-060, Recife, Pernambuco, Brazil. 3Departamento de Ciências Farmacêuticas, Universidade Federal da Paraíba, Campus I, Castelo Branco, 58051-970, Joao Pessoa, Paraíba, Brazil. Received 15 March, 2016; Accepted 20 May, 2016 Syagrus coronata (Mart.) Becc. (Arecaceae) is a native Brazilian palm (ouricuri) and despite the use of its derived products by traditional communities, few scientific reports have been published regarding its biomedical activity. This study investigates the chemical composition and anti-Staphylococcus aureus effects of both manufactured oil (SCO) and essential oil (SCEO) from S. coronata seeds. SCO was provided by rural inhabitants, while SCEO was obtained by hydrodistillation. Chemical characterization was performed by gas chromatography-mass spectrometry (GC/MS). In vitro antimicrobial activity was determined against 17 S. aureus strains, including multidrug-resistant strains. -

Analysing the History of the Derelomine Flower Weevil-Carludovica Association (Coleóptera: Curculionidae; Cyelanthaeeae)
Biological Journal of the Linnean Society, 2004, 81, 483•517. With 17 figures Analysing the history of the derelomine flower weevil-Carludovica association (Coleóptera: Curculionidae; Cyelanthaeeae) NICO M. FRANZ* Escuela de Biología, Universidad de Costa Rica, Ciudad Universitaria 'Rodrigo Fació', Costa Rica Received 14 April 2003; accepted for publication 23 September 2003 The evolutionary history of the interaction among species of derelomine flower weevils (Coleóptera: Curculionidae: Derelomini) and the Panama-hat palm Carludovica (Cyelanthaeeae) is analysed with emphasis on the congruence of (1) topologies and (2) character state transformations in each of the Neotropical clades. For this purpose cladistic analyses are complemented with host plant records, natural history information and selected morphological studies of the associated taxa. The interaction is specialized, involving pollination, oviposition into the inflorescences and the prédation of seeds (particularly within Systenotelus). As results from a range of standard coevolutionary methods of analysis indicate, however, events of colonization, extinction and independent (non-reciprocal) speciation have been abundant throughout the history of the association. At the same time it is possible to specify the homology and suc- cession of characters among species of derelomines and Carludovica and interpret them as reciprocal adaptations to attack and protect the seeds, respectively. It is argued that • in light of the limited evolutionary stability of many insect•plant interactions • the question of coevolution is most effectively addressed by combining information from the character- and topology-based approaches. © 2004 The Linnean Society of London, Biological Journal of the Linnean Society, 2004, 81, 483-517. ADDITIONAL KEYWORDS: cladistics • congruence • coevolution • Derelomini • Ganglionus • Perelleschus • pollination • reciprocal adaptation • seed prédation • Systenotelus. -

Interacciones Mutualistas. X. Antropocoria: Florestas Neotropicales Publicacions Del CRBA
Interacciones mutualistas. X. Antropocoria: florestas neotropicales Publicacions del CRBA Interacciones mutualistas entre animales y plantas X. Antropocoria: florestas neotropicales Juan Carlos Guix Coordinador del Proyecto Neopangea e-mail: [email protected] 1 © Centre de Recursos de Biodiversitat Animal, Facultat de Biologia, Universitat de Barcelona. 2021. Maig, 2021 Publicat per: Centre de Recursos de Biodiversitat Animal Facultat de Biologia Universitat de Barcelona Avinguda Diagonal 643 08028 Barcelona Spain [email protected] www.ub.edu/crba Guix, J.C. 2021. Interacciones mutualistas entre animales y plantas. X. Antropocoria: florestas neotropicales. Publicacions del Centre de Recursos de Biodiversitat Animal. Universitat de Barcelona, Volum 16, 107 pp. Portada: Las pluvisilvas situadas en las laderas orientales de los Andes se encuentran entre los bosques tropicales más diversos del mundo. Provincia de Napo, Ecuador. Foto: Juan Carlos Guix. 2 Interacciones mutualistas. X. Antropocoria: florestas neotropicales Publicacions del CRBA Interacciones mutualistas entre animales y plantas X. Antropocoria: florestas neotropicales Juan Carlos Guix Con frecuencia se asocia la floresta amazónica u otros tipos de selvas tropicales con la noción de una “naturaleza virgen”. Sin embargo, las florestas tropicales y subtropicales del Neotrópico están habitadas desde hace más de 14.000 años por pueblos indígenas, y estos a su vez han producido cambios apreciables en las comunidades de plantas de estos ecosistemas. La diseminación de semillas grandes en florestas neotropicales Los humanos y las redes de interacciones animal-planta De un modo general, las interacciones entre los humanos modernos (Homo sapiens) y los sistemas de diseminación de semillas han sido expresadas en forma de impactos antropogénicos relacionados con la fragmentación de hábitats, la sobrecaza y la extinción, a escala local o regional, de frugívoros de medio y gran porte (cf. -

MORPHOLOGY of FRUITS, DIASPORES, SEEDS, SEEDLINGS, and SAPLINGS of Syagrus Coronata (Mart.) Becc
652 Original Article MORPHOLOGY OF FRUITS, DIASPORES, SEEDS, SEEDLINGS, AND SAPLINGS OF Syagrus coronata (Mart.) Becc. MORFOLOGIA DE FRUTOS, DIÁSPOROS, SEMENTES, PLÂNTULAS E MUDAS DE Syagrus coronata (Mart.) Becc Sueli da Silva SANTOS-MOURA 1; Edilma Pereira GONÇALVES 2; Luan Danilo Ferreira de Andrade MELO 1; Larissa Guimarães PAIVA 1; Tatiana Maria da SILVA 1 1. Master's in Agricultural Production by the Rural Federal University of Pernambuco, Academic Unit of Garanhuns, Garanhuns, PE, Brazil; 2. Teacher, doctor at the Federal Rural University of Pernambuco, Academic Unit of Garanhuns, Garanhuns, PE, Brazil. ABSTRACT: Licuri ( Syagrus coronata (Mart.) Becc.) is an ornamental palm tree native of Brazil with great economic potential, because it provides raw material for manufacturing a wide range of products. The objective of this study was to assess the morphology of the fruits, diaspores, seeds, seedlings, and saplings of Syagrus coronata . The study was performed at the Laboratory of Seed Analysis (LSA) of the Federal Rural University of Pernambuco/Academic Unit of Garanhuns-PE, by using licuri fruits collected from the rural area of Caetés-PE. It was evaluated fruit morphology, diaspores, seeds, seedlings and saplings. Germination, in the form of cotyledon petiole emergence, began 15 days after sowing, is hypogeal, cryptocotylar, and remote tubular. It is slow and uneven, extending up to 60 days after the first eophyll appears. The saplings have alternate, pinnate, glabrous, entire leaves with parallel venation and sheath invagination. The primary roots persistent, the secondary roots arise from the stem root node in the primary root, and lateral roots only fasciculate was evidenced when the change was 300 days, and must remain in the nursery for at least 360 days after germination before taking it to the field, due to the slow development of this species. -

Sfps Fall 2011 Sale Plant List
SFPS FALL 2011 SALE PLANT LIST PLANTS VENDOR # Palms Acanthophoenix rubra 35 Acoelorrhaphe wrightii 26, 67 Acrocomia aculeata 50, 67 Actinokentia divaricata 35, 57, 66, 68, 72 Actinorhytis calapparia 72 Adonidia merrillii 31, 57, 66, 89 Adonidia merrillii var. "Golden Form" 35 Aiphanes aculeata = Aiphanes horrida - Aiphanes caryotifolia = Aiphanes horrida - Aiphanes erosa = Aiphanes minima - Aiphanes horrida 35, 68, 72 Aiphanes minima 68 Aiphanes vincentiana = Aiphanes minima - Allagoptera arenaria 57, 66, 67, 68, 72 Allagoptera campestris 67 Allagoptera leucocalyx 57 Alloschmidia glabrata = Basselinia glabrata - Alsmithia longipes = Heterospathe longipes - Archontophoenix cunninghamiana var. 'Illawara' 68 Archontophoenix maxima 67, 72 Archontophoenix myolensis 50, 66, 67, 68 Archontophoenix purpurea 57, 66, 72 Archontophoenix tuckeri 66, 68 Areca aliceae = Areca triandra - Areca camarinensis 57, 68 Areca catechu 57, 67, 72 Areca catechu var. 'Dwarf' 35, 50 Areca hutchinsoniana 68 Areca ipot 67 Areca latiloba = Areca montana - Areca macrocalyx var. 'Red Form' 35, 57, 68 Areca macrocarpa 68 Areca montana 57 Areca triandra 68, 72 Areca vestiaria 25, 35, 57, 67, 68 Areca vestiaria var. 'Orange Form' 25, 57, 67, 72 Areca vestiaria var. 'Maroon Leaf' 35, 57, 67 Areca vestiaria var. 'Red Leaf' 57, 67, 72 Areca sp. 'Yellow Crownshaft' 25 Arenga ambong = Arenga undulatifolia - Arenga brevipes 57 Arenga caudata 66 Arenga engleri 31, 66, 68, 72 Arenga hookeriana 35, 57, 66, 72 Arenga microcarpa 26, 66 Arenga obtusifolia 57, 66 PLANTS VENDOR # Arenga pinnata 50, 57, 66, 67, 68 Arenga porphyrocarpa 66 Arenga tremula 26, 57, 66, 68, 72 Arenga undulatifolia 35, 57, 66, 67 Arenga westerhoutii 68 Asterogyne martiana 57, 68, 72 Astrocaryum acaule 72 Astrocaryum alatum 35, 50, 57, 67 Astrocaryum mexicanum 72 Astrocaryum murumuru 72 Attalea butyracea 57, 67, 72 Attalea cohune 35 Attalea phalerata 50, 91 Attalea rostrata 68 Attalea speciosa 50, 66 Bactris bidentula 72 Bactris gasipaes 67 Bactris gasipaes var. -

3.7.10 Curculioninae Latreille, 1802 Jetzt Beschriebenen Palaearctischen Ceuthor- Rhynchinen
Curculioninae Latreille, 1802 305 Schultze, A. (1902): Kritisches Verzeichniss der bis 3.7.10 Curculioninae Latreille, 1802 jetzt beschriebenen palaearctischen Ceuthor- rhynchinen. – Deutsche Entomologische Zeitschrift Roberto Caldara , Nico M. Franz, and Rolf 1902: 193 – 226. G. Oberprieler Schwarz, E. A. (1894): A “ parasitic ” scolytid. – Pro- ceedings of the Entomological Society of Washington 3: Distribution. The subfamily as here composed (see 15 – 17. Phylogeny and Taxonomy below) includes approx- Scudder, S. H. (1893): Tertiary Rhynchophorous Coleo- ptera of the United States. xii + 206 pp. US Geological imately 350 genera and 4500 species (O ’ Brien & Survey, Washington, DC. Wibmer 1978; Thompson 1992; Alonso-Zarazaga Stierlin, G. (1886): Fauna insectorum Helvetiae. Coleo- & Lyal 1999; Oberprieler et al. 2007), provisionally ptera helvetiae , Volume 2. 662 pp. Rothermel & Cie., divided into 34 tribes. These are geographically Schaffhausen. generally restricted to a lesser or larger degree, only Thompson, R. T. (1973): Preliminary studies on the two – Curculionini and Rhamphini – being virtually taxonomy and distribution of the melon weevil, cosmopolitan in distribution and Anthonomini , Acythopeus curvirostris (Boheman) (including Baris and Tychiini only absent from the Australo-Pacifi c granulipennis (Tournier)) (Coleoptera, Curculion- region. Acalyptini , Cionini , Ellescini , Mecinini , idae). – Bulletin of Entomological Research 63: 31 – 48. and Smicronychini occur mainly in the Old World, – (1992): Observations on the morphology and clas- from Africa to the Palaearctic and Oriental regions, sifi cation of weevils (Coleoptera, Curculionidae) with Ellescini, Acalyptini, and Smicronychini also with a key to major groups. – Journal of Natural His- extending into the Nearctic region and at least tory 26: 835 – 891. the latter two also into the Australian one. -

Record of Tritrophic Relationship Between Syagrus Coronata (Martius)
doi: 10.12741/ebrasilis.v14.e922 e-ISSN 1983-0572 Creative Commons License v4.0 (CC-BY) Copyright © Author(s) Article Full Open Access Scientific Note Record of tritrophic relationship between Syagrus coronata (Martius) Beccari (Arecaceae), Pachymerus nucleorum Fabricius (Coleoptera: Chrysomelidae: Bruchinae) and Heterospilus sp. (Hymenoptera: Braconidae) in the State of Alagoas, northeastern Brazil Jefferson Duarte de Melo , Suianne Oliveira dos Santos Cajé & Iracilda Maria de Moura Lima Laboratório de Bioecologia de Insetos, Instituto de Ciências Biológicas e da Saúde, Universidade Federal de Alagoas, Maceió, Alagoas, Brazil. EntomoBrasilis 14: e922 (2021) Edited by: Abstract. Some conservation units in Brazil border urban areas, like the Catolé and Fernão Velho William Costa Rodrigues Environmental Protection Area (EPA) in the State of Alagoas. In urban areas, there is the habit of cultivating plants for landscape purposes, and Syagrus coronata (Martius) Beccari (Arecaceae), “Licuri” Article History: or “Ouricuri”, is a palm tree commonly used in ornamentation; a native species from Caatinga and Received: 23.vii.2020 Atlantic Forest biomes widely explored through time. Some insects have part of their development Accepted: 30.xii.2020 associated with plants, and Pachymerus nucleorum Fabricius (Coleoptera: Chrysomelidae: Bruchinae) Published: 06.iii.2021 has a close connection with some Arecaceae. Females usually lay eggs on the surface of fallen fruits and the immatures feed on the seed under the drupe endocarp; the larvae, even protected by the Corresponding author: hard surface could be preyed by skilled parasitoid wasps. Here, the record of a tritrophic relationship Jefferson Duarte de Melo between S. coronata, P. nucleorum, and a wasp of the genus Heterospilus (Hymenoptera: Braconidae) [email protected] in an urbanized region of Alagoas, close to a remnant of Atlantic Forest of the Catolé and Fernão Funding agencies: Velho EPA is communicated.