Clonal Spread in Second Growth Stands of Coast Redwood, Sequoia Sempervirens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Austrocedrus Forests of South America Are Pivotal Ecosystems at Risk Due to the Emergence of an Exotic Tree Disease
GSDR 2015 Brief Austrocedrus forests of South America are pivotal ecosystems at risk due to the emergence of an exotic tree disease: can a joint effort of research and policy save them? By Alina Greslebin 1, Maria Laura Vélez 2 and Matteo Garbelotto 3. 1CONICET-Universidad Nacional de la Patagonia SJB, Argentina; 2CONICET-Centro de Investigación y Extensión Forestal Andino Patagónico (CIEFAP), Argentina; 3University of California Berkeley, USA Introduction A. chilensis covers today a total estimated area of Human expansion, global movement, and climate 185,000 ha in South America. As a dominant forest change have led to a number of emerging and re- species, its role in supporting biodiversity, generating emerging diseases. The decline of biodiversity due to shelter for wildlife, as well as preventing soil erosion emerging plants pathogens may cause habitat and and preserving water quality is well understood. Along wildlife loss and declines in ecosystem services. This, in with Araucaria araucana, it is the tree species that turn, often results in lower human well-being. Reports grows furthest into the ecotone zone within the of emerging plant diseases are constantly on the rise, Patagonia steppe, where it plays a key role preventing and often they appear to be linked to the commercial desertification. There are however additional functions trade of plants and plant products. While there are this tree provides, including the production of valuable several examples of decimation or extinction of plant timber and the generation of an environment ideal for hosts affected by invasive forest diseases, there are no cattle grazing, recreational and touristic activities and known cases of invasive forest diseases successfully for human settlement. -

Supporting Information
Supporting Information Mao et al. 10.1073/pnas.1114319109 SI Text BEAST Analyses. In addition to a BEAST analysis that used uniform Selection of Fossil Taxa and Their Phylogenetic Positions. The in- prior distributions for all calibrations (run 1; 144-taxon dataset, tegration of fossil calibrations is the most critical step in molecular calibrations as in Table S4), we performed eight additional dating (1, 2). We only used the fossil taxa with ovulate cones that analyses to explore factors affecting estimates of divergence could be assigned unambiguously to the extant groups (Table S4). time (Fig. S3). The exact phylogenetic position of fossils used to calibrate the First, to test the effect of calibration point P, which is close to molecular clocks was determined using the total-evidence analy- the root node and is the only functional hard maximum constraint ses (following refs. 3−5). Cordaixylon iowensis was not included in in BEAST runs using uniform priors, we carried out three runs the analyses because its assignment to the crown Acrogymno- with calibrations A through O (Table S4), and calibration P set to spermae already is supported by previous cladistic analyses (also [306.2, 351.7] (run 2), [306.2, 336.5] (run 3), and [306.2, 321.4] using the total-evidence approach) (6). Two data matrices were (run 4). The age estimates obtained in runs 2, 3, and 4 largely compiled. Matrix A comprised Ginkgo biloba, 12 living repre- overlapped with those from run 1 (Fig. S3). Second, we carried out two runs with different subsets of sentatives from each conifer family, and three fossils taxa related fi to Pinaceae and Araucariaceae (16 taxa in total; Fig. -

Methods for Measuring Frost Tolerance of Conifers: a Systematic Map
Review Methods for Measuring Frost Tolerance of Conifers: A Systematic Map Anastasia-Ainhoa Atucha Zamkova *, Katherine A. Steele and Andrew R. Smith School of Natural Sciences, Bangor University, Bangor LL57 2UW, Gwynedd, UK; [email protected] (K.A.S.); [email protected] (A.R.S.) * Correspondence: [email protected] Abstract: Frost tolerance is the ability of plants to withstand freezing temperatures without unrecov- erable damage. Measuring frost tolerance involves various steps, each of which will vary depending on the objectives of the study. This systematic map takes an overall view of the literature that uses frost tolerance measuring techniques in gymnosperms, focusing mainly on conifers. Many different techniques have been used for testing, and there has been little change in methodology since 2000. The gold standard remains the field observation study, which, due to its cost, is frequently substituted by other techniques. Closed enclosure freezing tests (all non-field freezing tests) are done using various types of equipment for inducing artificial freezing. An examination of the literature indicates that several factors have to be controlled in order to measure frost tolerance in a manner similar to observation in a field study. Equipment that allows controlling the freezing rate, frost exposure time and thawing rate would obtain results closer to field studies. Other important factors in study design are the number of test temperatures used, the range of temperatures selected and the decrements between the temperatures, which should be selected based on expected frost tolerance of the tissue and species. Citation: Atucha Zamkova, A.-A.; Steele, K.A.; Smith, A.R. -

Taxodium Distichum
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School June 2018 Assessing Bald Cypress (Taxodium distichum) Tree Dynamic Change in USF Forest Preserve Area Using Mixture-Tuned Matched Filtering and Multitemporal Satellite Imagery Yujia Wang University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Geology Commons Scholar Commons Citation Wang, Yujia, "Assessing Bald Cypress (Taxodium distichum) Tree Dynamic Change in USF Forest Preserve Area Using Mixture- Tuned Matched Filtering and Multitemporal Satellite Imagery" (2018). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/7375 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Assessing Bald Cypress (Taxodium distichum) Tree Dynamic Change in USF Forest Preserve Area Using Mixture-Tuned Matched Filtering and Multitemporal Satellite Imagery by Yujia Wang A thesis submitted in partial fulfillment of the requirement for the degree of Master of Science School of Geosciences College of Art and Science University of South Florida Co-Major Professor: Ping Wang, Ph.D. Co-Major Professor: Ruiliang Pu, Ph.D. Joni Downs, Ph.D. Date of Approval: June 26, 2018 Keywords: Remote Sensing, bald cypress tree, Linear Spectral Unmixing, abundance -

Temperate Coniferous Forests
Temperate Coniferous Forests Richard H Waring Volume 2, The Earth system: biological and ecological dimensions of global environmental change, pp 560–565 Edited by Professor Harold A Mooney and Dr Josep G Canadell in Encyclopedia of Global Environmental Change (ISBN 0-471-97796-9) Editor-in-Chief Ted Munn John Wiley & Sons, Ltd, Chichester, 2002 and sunken stomata and a large proportion of their stems Temperate Coniferous consists of sapwood that serves as a storage reservoir, as Forests well as a conduit for transport of water from roots to leaves. Richard H Waring GEOGRAPHIC DISTRIBUTION Oregon State University, Eugene, OR, USA Today, temperate evergreen coniferous forests cover app- roximately 2.4 ð 106 km2 (Melillo et al., 1993; Landsberg Temperate coniferous forests include representatives of the and Gower, 1997). Conifers dominate the montane forests most massive forms of terrestrial life on Earth. In the in North America, Europe, and China. Smaller areas of tem- coastal redwood (Sequoia sempervirens) forests of northern perate conifers are located in montane regions of Korea, California, trees may reach heights >100 m and accumu- Japan, Mexico, Nicaragua, and Guatemala. In Europe, the late biomass in excess of 3000 t ha1 . In the more extensive distribution of native and introduced conifers has been Douglas fir (Pseudotsuga menziesii) forests of the Pacific expanded into areas more climatically favorable to hard- Northwest, USA and Canada, biomass in older stands fre- woods. Pines (Pinus), represented by more than 90 species, quently reaches 1000 t ha1 , with an additional 500 t ha1 are the most widely distributed conifer and their range has of standing dead and fallen trees. -

Restoration of Burned and Post-Fire Logged Austrocedrus Chilensis Stands in Patagonia
SPECIAL ISSUE CSIRO PUBLISHING International Journal of Wildland Fire 2019, 28, 365–376 https://doi.org/10.1071/WF18154 Restoration of burned and post-fire logged Austrocedrus chilensis stands in Patagonia: effects of competition and environmental conditions on seedling survival and growth M. Florencia UrretavizcayaA,B,D and Guillermo E. Defosse´ A,C AConsejo Nacional de Investigaciones Cientı´ficas y Tecnolo´gicas (CONICET), Centro de Investigacio´n y Extensio´n Forestal Andino Patago´nico (CIEFAP), CC 14 Esquel (9200) Chubut, Argentina. BMinisterio de la Produccio´n, Provincia de Chubut, Argentina. CUniversidad Nacional de la Patagonia San Juan Bosco, Facultad de Ingenierı´a, Esquel, Chubut, Argentina. DCorresponding author. Email: [email protected] Abstract. In Andean Patagonia, Argentina, severe wildfires produce high mortality in Austrocedrus chilensis forests. Owing to its high timber quality, A. chilensis trees are generally salvage logged right after wildfires. Post-logged areas result in open, denuded stands with low herbaceous or shrub cover, which precludes natural A. chilensis regeneration. In severely burned A. chilensis stands, we determined how different site conditions (salvage logging, SL, and without logging, WL), and the combination of different methods of control of above- and belowground competition and micro- environmental factors (incident radiation, soil temperature and soil moisture) affected survival, growth and hydric status of planted A seedlings. Two growing seasons after plantation, seedling survival was lower than 10% at SL sites, whereas it was near 90% at WL sites. Four seasons after establishment, and regardless of competition treatments, no seedlings survived at SL sites, whereas over 75% survived at WL sites. Radiation attenuation by canopy of WL sites benefitted A. -

Published Version
Tree ings and ople International Conference on the Future of Dendrochronology Davos, 22-26 September 2001 Abstracts Edited by Michele Kaennel Dobbertin and Otto Ueli Braker Published by the Swiss Federal Research Institute WSL CH-8903 Birmensdorf. 2001 Recommended form of citation: Kaennel Dobbertin, M.; Braker, O.U. (eds) 2001: Tree Rings and People. International Conference on the Future of Dendrochronology. Davos, 22-26 September 2001. Abstracts. Birmensdorf, Swiss Federal Research Institute WSL. 278 pp. ISBN 3-905620-96-0 Available from Library WSL ZOrcherstrasse 111 CH-8903 Birmensdorf E-mail: [email protected] Contents In the table of contents, short and extended* poster abstracts are sorted according to sub-themes within each session. Extended abstracts are grouped in the last section of this volume, pp. 197-264. Session 1: Wood structure and function The increment puncher: A tool for extracting small cores of wood and bark from living trees 17 Theodor Forster, Fritz Hans Schweingruber, Bernhard Denneler Using three different methods for measuring tree rings: comparison of results 18 Ignacio Garcfa Gonzalez, Lorena Garcfa Rodrfguez, Elvira Dfaz Vizcafno A computer-assisted dendrochronology workstation: progress report 19 Martin A.R. Munro, Malcolm K. Hughes, Robert Schowengerdt, W. Steven Conner, James B. Engle, Giribalan Gopalan, James M. Burns Analysis of tree-ring growth and cell structure on free-hand sections by confocal microscopy * 197 Peter Kitin, Ryo Funada Microphotometry and tomography as tools for the study of internal wood structures 20 Vladimir Bahyl, J. Dursk, M. Gajtanska, Pham Van Tinh, Th. Seifert, A. Zbonak1 Image analysis of ring anatomy: interpretation of intra-ring microvariations in wood density * 198 Valerie Decoux, Emmanuel Defays, Charles De Canniere Modelling of cambial activity 21 Alexander V. -
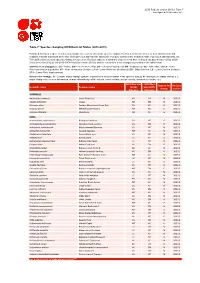
Table 7: Species Changing IUCN Red List Status (2012-2013)
IUCN Red List version 2013.2: Table 7 Last Updated: 25 November 2013 Table 7: Species changing IUCN Red List Status (2012-2013) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2012 (IUCN Red List version 2012.2) and 2013 (IUCN Red List version 2013.2) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.) IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2012) List (2013) change version Category Category MAMMALS Nycticebus javanicus Javan Slow Loris EN CR N 2013.2 Okapia johnstoni Okapi NT EN N 2013.2 Pteropus niger Greater Mascarene Flying -

Research Report South American Chronology
Fitzroya Cupressoides Yields 1534- Year Long South American Chronology Item Type Article Authors Boninsegna, José A.; Holmes, Richard L. Citation Boninsegna, J.A., Holmes, R.L. 1985. Fitzroya cupressoides yields 1534-year long South American chronology. Tree-Ring Bulletin 45:37-42. Publisher Tree-Ring Society Journal Tree-Ring Bulletin Rights Copyright © Tree-Ring Society. All rights reserved. Download date 29/09/2021 16:30:06 Link to Item http://hdl.handle.net/10150/261336 TREE -RING BULLETIN, Vol. 45, 1985 RESEARCH REPORT FITZROYA CUPRESSOIDES YIELDS 1534 -YEAR LONG SOUTH AMERICAN CHRONOLOGY JOSÉ A. BONINSEGNA Instituto Argentino de Nivolo gia y Glaciología Mendoza - Argentina and RICHARD L. HOLMES Laboratory of Tree -Ring Research University of Arizona Tucson, Arizona ABSTRACT The longest tree -ring chronologies for the Southern Hemisphere published to date go back to A.D. 1011 in central Chile; 1028 in Tasmania, Australia; 1140 in western Argentina; and 1256 on the North Island, New Zealand. For paleoclimatic and other studies longer time series would be very desirable. Here we report on the first successful crossdating and chronology development for Fitzroya cupressoides,a redwood -like conifer in western Argentina, which goes back to 441 and exhibits desirable statistical characteristics. INTRODUCTION Fitzroya cupressoides (Molina (Johnson)), common name "alerce," is a tall conifer which grows in the subantarctic forest of Argentina and Chile. The species is long - lived, a highly desirable characteristic in the development of tree -ring chronologies. Increment core samples of Fitzroya cupressoides were collected from adjacent sites by Schulman in 1949, Lerman in 1962, and by a group of us under the direction of LaMarche, in 1975. -

Phytophthora Austrocedrae
Greslebin, A., Hansen, E. M., and La Manna, L. 2011. Phytophthora austrocedrae. Forest s”, a lethal disease of Austrocedrus in Argentina. Austrocedrus chilensis Phytophthoras 1(1). doi: 10.5399/osu/fp.1.1.1806 Phytophthora austrocedrae Overview Phytophthora austrocedrae Gresl. & E.M. Hansen (2007) was isolated from necrotic lesions of stem and roots of Austrocedrus chilensis (Cupressaceae). It is homothallic with semi-papillate sporangia, oogonia with Sporangium with distorted shape, from Q-bank (left); morphology of sporangia from Greslebin et al. 2007 (right) used with permission. Genetics The ITS sequence was identical to sequences of Phytophthora DNA extracted from bark of diseased trees. P. austrocedrae is in clade 8 of the Cooke et al. (2000) molecular phylogeny of the genus, with P. lateralis, another pathogen of Cupressaceae, and other aggressive pathogenic species. P. syringae is the closest described relative. Phylogeny of P. austrocedrae based on ITS rDNA sequence analysis (Greslebin et al. 2007)<> Growth In V8 and tomato juice (TA) agar the colony was uniform, without growth pattern, cottony, dome-shaped in the center and appressed or mostly submerged at the margins. In corn meal agar (CMA) the colony was appressed, with little or no aerial mycelium and the submerged mycelia showed an arachnoid pattern. In potato dextrose agar (PDA) the colony was uniform, without growth pattern, densely felty to woolly, with abundant and dense aerial mycelium. Growth was very slow and favored by cool temperatures. Optimum temperature was 17.5°C, with no growth at 25°C. Maximum radial growth rate at optimum temperature on V8 agar ranged from 1.0 – 1.8 mm/day. -

Sequoiadendron Giganteum)
Article Patterns of Fine-Scale Spatial Genetic Structure and Pollen Dispersal in Giant Sequoia (Sequoiadendron giganteum) Rainbow DeSilva * and Richard S. Dodd Deptartment of Environmental Science, Policy, and Management, University of California, Berkeley, CA 94720, USA; [email protected] * Correspondence: [email protected] Abstract: Research Highlights: Patterns of dispersal shape the distribution and temporal devel- opment of genetic diversity both within and among populations. In an era of unprecedented environmental change, the maintenance of extant genetic diversity is crucial to population persis- tence. Background and Objectives: We investigate patterns of pollen dispersal and spatial genetic structure within populations of giant sequoia (Sequoiadendron giganteum). Materials and Methods: The leaf genotypes of established trees from twelve populations were used to estimate the extent of spatial genetic structure within populations, as measured by the Sp statistic. We utilized progeny arrays from five populations to estimate mating parameters, the diversity of the pollen pool, and characteristics of pollen dispersal. Results: Our research indicates that giant sequoia is predom- inantly outcrossing, but exhibits moderate levels of bi-parental inbreeding (0.155). The diversity of the pollen pool is low, with an average of 7.5 pollen donors per mother tree. As revealed by the Sp-statistic, we find significant genetic structure in ten of twelve populations examined, which indicates the clustering of related individuals at fine spatial scales. Estimates of pollen and gene dispersal indicate predominantly local dispersal, with the majority of pollen dispersal <253 m, and with some populations showing fat-tailed dispersal curves, suggesting potential for long-distance dispersal. Conclusions: The research presented here represent the first detailed examination of the reproductive ecology of giant sequoia, which will provide necessary background information for the Citation: DeSilva, R.; Dodd, R.S. -

Vegetative Propagation of Patagonian Cypress, a Vulnerable Species from the Subantarctic Forest of South America
BOSQUE 30(1): 18-26, 2009 Vegetative propagation of patagonian cypress, a vulnerable species from the subantarctic forest of South America Propagación vegetativa del ciprés de la cordillera, una especie vulnerable del bosque subantártico de Sudamérica Alejandro Aparicioa*, Mario Pastorinoa, b, Alejandro Martinez-Meiera, Leonardo Galloa *Corresponding author: aInstituto Nacional de Tecnología Agropecuaria, Unidad de Genética Forestal, EEA Bariloche, CC 277 (8400) Bariloche, Río Negro, Argentina, tel.: +54 02944 422731, fax: +54 02944 424991, [email protected] bCONICET, Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina. SUMMARY Increasing risks of loosing forest genetic resources and the need for a sustainable quality wood supply make focusing research efforts on undomesticated tree species necessary. The study of their genetic variation in vegetative propagation ability and the development of practical methodologies to obtain genetic copies are essential tools for conservation and breeding programs. patagonian cypress Austrocedrus chilensis is a vulnerable conifer native to the subantarctic forest of South America, with high ecological importance and economical potential. Its genetic diversity, mostly sheltered within unprotected populations, is seriously threatened. This work examined the ability of different genotypes of A. chilensis from xeric, mesic and humid populations to propagate asexually by stem rooting and scion grafting. The influence of exogenous auxins, season of cutting collection, ontogenetic stage of the donor plants, grafting season, and type of scions, was analyzed. Stem rooting ability was found to be poor both for adult (average = 0.27%) and juvenile ortets (average = 2.10%). Only one treatment, a juvenile progeny from a xeric provenance, showed remarkable results with 33.33% of rooting capacity.