Nested Radiations and the Pulse of Angiosperm Diversification
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Visual Guide to Collecting Plant Tissues for DNA
A visual guide to collecting plant tissues for DNA Collecting kit checklist Silica gel1 Permanent marker and pencil Resealable bags, airtight plastic container Razor blade / Surgical scissors Empty tea bags or coffee filters Ethanol and paper tissue or ethanol wipes Tags or jewellers tags Plant press and collecting book 1. Selection and preparation of fresh plant tissue: Sampling avoided. Breaking up leaf material will bruise the plant tissue, which will result in enzymes being released From a single plant, harvest 3 – 5 mature leaves, or that cause DNA degradation. Ideally, leaf material sample a piece of a leaf, if large (Picture A). Ideally should be cut into smaller fragments with thick a leaf area of 5 – 10 cm2 should be enough, but this midribs being removed (Picture C). If sampling robust amount should be adjusted if the plant material is leaf tissue (e.g. cycads, palms), use a razor blade or rich in water (e.g. a succulent plant). If leaves are surgical scissors (Picture D). small (e.g. ericoid leaves), sample enough material to equate a leaf area of 5 – 10 cm2. If no leaves are Succulent plants available, other parts can be sampled such as leaf buds, flowers, bracts, seeds or even fresh bark. If the If the leaves are succulent, use a razor blade to plant is small, select the biggest specimen, but never remove epidermal slices or scoop out parenchyma combine tissues from different individuals. tissue (Picture E). Cleaning Ideally, collect clean fresh tissues, however if the leaf or plant material is dirty or shows potential contamination (e.g. -

Chloroplast Genome Analysis of Angiosperms and Phylogenetic Relationships Among
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Chloroplast genome analysis of Angiosperms and phylogenetic relationships among Lamiaceae members with particular reference to teak (Tectona grandis L.f) P. MAHESWARI, C. KUNHIKANNAN AND R. YASODHA* Institute of Forest Genetics and Tree Breeding, Coimbatore 641 002 INDIA *Author for correspondence R. YASODHA, Institute of Forest Genetics and Tree Breeding, Coimbatore, India Telephone: +91 422 2484114; Fax number : +91 422 248549; e.mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Availability of comprehensive phylogenetic tree for flowering plants which includes many of the economically important crops and trees is one of the essential requirements of plant biologists for diverse applications. It is the first study on the use of chloroplast genome of 3265 Angiosperm taxa to identify evolutionary relationships among the plant species. Sixty genes from chloroplast genome was concatenated and utilized to generate the phylogenetic tree. Overall the phylogeny was in correspondence with Angiosperm Phylogeny Group (APG) IV classification with very few taxa occupying incongruous position either due to ambiguous taxonomy or incorrect identification. Simple sequence repeats (SSRs) were identified from almost all the taxa indicating the possibility of their use in various genetic analyses. -
BM CC EB What Can We Learn from a Tree?
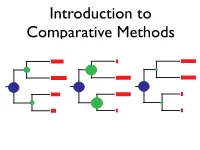
Introduction to Comparative Methods BM CC EB What can we learn from a tree? Net diversification (r) Relative extinction (ε) Peridiscaceae Peridiscaceae yllaceae yllaceae h h atop atop Proteaceae Proteaceae r r Ce Ce Tr oc T ho r M M o de c y y H H C C h r r e e a D D a o o nd o e e G G a a m m t t a a d r P r P h h e e u u c c p A p A r e a a e e a a a c a c n n a i B i B h h n d m d l m a l a m m e a e a e e t t n n c u u n n i i d i i e e e e o n o n p n p n a e a S e e S e e n n x x i i r c c a a n o n o p p h g e h g ae e l r a l r a a a a a a i i a a a e a e i b i b h y d c h d c i y i c a a c x c x c c G I a G I a n c n c c c y l y l t a a t a a e e e e e i l c i l c m l m l e c e c f a e a a f a e a a l r r l c c a i i r l e t e t a a r l a a e e u u u u o a o a a a c a c a a l a l e e e b b a a a a e e c e e c a a s c s c c e l c e l e e g e g e a a a a e e n n s e e s e e e e a a a P a P e e N N u u u S u S a e a e a a e e c c l n a l n e e a e e a e a e a e a e r a r a c c C i C i R R a e a e a e a e r c r c A A a d a a d a e i e i phanopetalaceae s r e ph s r e a a s e c s e c e e u u b a a b a e e P P r r l l e n e a a a a m m entho e e e Ha a H o a c r e c r e nt B B e p e e e c e e c a c c h e a p a a p a lo lo l l a a e s o t e s i a r a i a r r r r r a n e a n e a b a l b t a t gaceae e g e ceae a c a s c s a z e z M i a e M i a c a d e a d e ae e ae r e r a e a e a a a c c ce r e r L L i i ac a Vitaceae Vi r r C C e e ta v e v e a a c a a e ea p e ap c a c a e a P P e e l l e Ge G e e ae a t t e e p p r r ce c an u an -

Supplementary Information
Supporting Information for Cornwell et al. 2014. Functional distinctiveness of major 1 plant lineages. doi: 10.1111/1365-2745.12208. SUPPORTING INFORMATION Summary In this Supporting Information, we report additional methods details with respect to the trait data, the climate data, and the analysis. In addition, to more fully explain the method, we include several additional figures. We show the top-five lineages, including the population of lineages from which they were selected, for five important traits (Figure S1 on five separate pages); the bivariate distribution of three selected clades with respect to SLA and leaf N, components of the leaf economic spectrum (Figure S2), the geographic distribution of the clades where this proved useful for interpretation (Figure S3); the procedure for selecting the top six nodes in the leaf N trait, to illustrate the internal behavior of our new comparative method, especially the relative contribution of components of extremeness and the sample size weighting to the results (Figure S4). Finally, we provide references for data used in analyses. SUPPORTING METHODS TRAITS DATABASE We compiled a database for five plant functional traits. Each of these traits is the result of a separate research initiative in which data were gathered directly from researchers leading those efforts and/or the literature; in most, but not all cases, these data have been published elsewhere. Detailed methods for data collection and assembly for each trait are available in the original publications; further data were added for some traits from the primary literature (for references see description of individual traits and Supporting References below). For our compilation, all data were brought to common units for a given trait and thoroughly error checked. -

2 ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM History Of
ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM The Angiosperm Phylogeny Group, or APG, refers to an informal international group of systematic botanists who came together to try to establish a consensus view of the taxonomy of flowering plants (angiosperms) that would reflect new knowledge about their relationships based upon phylogenetic studies. As of 2010, three incremental versions of a classification system have resulted from this collaboration (published in 1998, 2003 and 2009). An important motivation for the group was what they viewed as deficiencies in prior angiosperm classifications, which were not based on monophyletic groups (i.e. groups consisting of all the descendants of a common ancestor). APG publications are increasingly influential, with a number of major herbaria changing the arrangement of their collections to match the latest APG system. Angiosperm classification and the APG Until detailed genetic evidence became available, the classification of flowering plants (also known as angiosperms, Angiospermae, Anthophyta or Magnoliophyta) was based on their morphology (particularly that of the flower) and their biochemistry (what kinds of chemical compound they contained or produced). Classification systems were typically produced by an individual botanist or by a small group. The result was a large number of such systems (see List of systems of plant taxonomy). Different systems and their updates tended to be favoured in different countries; e.g. the Engler system in continental Europe; the Bentham & Hooker system in Britain (particularly influential because it was used by Kew); the Takhtajan system in the former Soviet Union and countries within its sphere of influence; and the Cronquist system in the United States. -

The Woody Planet: from Past Triumph to Manmade Decline
plants Review The Woody Planet: From Past Triumph to Manmade Decline Laurence Fazan 1, Yi-Gang Song 2,3 and Gregor Kozlowski 1,3,4,* 1 Department of Biology and Botanical Garden, University of Fribourg, Chemin du Musée 10, 1700 Fribourg, Switzerland; [email protected] 2 Eastern China Conservation Center for Wild Endangered Plant Resources, Shanghai Chenshan Botanical Garden, Chenhua Road No.3888, Songjiang, Shanghai 201602, China; [email protected] 3 Shanghai Chenshan Plant Science Research Center, Chinese Academy of Sciences, Chenhua Road No.3888, Songjiang, Shanghai 201602, China 4 Natural History Museum Fribourg, Chemin du Musée 6, 1700 Fribourg, Switzerland * Correspondence: [email protected]; Tel.: +41-26-300-88-42 Received: 6 November 2020; Accepted: 16 November 2020; Published: 17 November 2020 Abstract: Woodiness evolved in land plants approximately 400 Mya, and very soon after this evolutionary invention, enormous terrestrial surfaces on Earth were covered by dense and luxurious forests. Forests store close to 80% of the biosphere’s biomass, and more than 60% of the global biomass is made of wood (trunks, branches and roots). Among the total number of ca. 374,000 plant species worldwide, approximately 45% (138,500) are woody species—e.g., trees, shrubs or lianas. Furthermore, among all 453 described vascular plant families, 191 are entirely woody (42%). However, recent estimations demonstrate that the woody domination of our planet was even greater before the development of human civilization: 1.4 trillion trees, comprising more than 45% of forest biomass, and 35% of forest cover disappeared during the last few thousands of years of human dominance on our planet. -

DDC) Stemming from the Adoption of the APG (Angiosperm Phylogeny Group) III Classification As the Basis for the DDC’S Treatment of Flowering Plants
This PDF documents proposed changes throughout the Dewey Decimal Classification (DDC) stemming from the adoption of the APG (Angiosperm Phylogeny Group) III classification as the basis for the DDC’s treatment of flowering plants. We request comment from any interested party, to be sent to Rebecca Green ([email protected]) by 31 January 2016. Please include “Angiosperm review comments” in your subject line. -------------------------------------------------------------- Why is the DDC adopting a new basis for classifying angiosperms (flowering plants)? During the latter half of the 20th century, biological classification turned from establishing taxa predominantly on the basis of morphological similarities to establishing taxa predominantly on the basis of shared ancestry / shared derived characters, with biological taxonomies mirroring evolutionary relationships. Phylogenetic analysis typically underlies modern evolutionary classifications, but has resulted in the development of many competing classifications. Within the domain of flowering plants, different classification systems have been favored in different countries. The Angiosperm Phylogeny Group, a global consortium of botanists, has addressed this issue by developing a “consensus” classification that is monophyletic (i.e., its taxa include all but only the descendants of a common ancestor). Now in its third version, the APG III classification is considered relatively stable and useful for both research and practice (e.g., for organizing plants in herbaria). The development for flowering plants presented here is the culmination of DDC editorial work over a span of several years. An early version revised 583–584 to make the schedule compatible with the APG III classification, while trying to minimize relocations and using see references to establish the APG III logical hierarchy. -

Advances in Botanical Research, Volume 54
Advances in BOTANICAL RESEARCH Series Editors JEAN-CLAUDE KADER Laboratoire Physiologie Cellulaire et Mole´culaire des Plantes, CNRS, Universite´ de Paris, Paris, France MICHEL DELSENY Laboratoire Ge´nome et De´veloppement des Plantes, CNRS IRD UP, Universite´ de Perpignan, Perpignan, France Advances in BOTANICAL RESEARCH Series Editors JEAN-CLAUDE KADER Laboratoire Physiologie Cellulaire, et Mole´culaire des Plantes, CNRS, Universite´ de Paris, Paris, France MICHEL DELSENY Laboratoire Ge´nome et De´veloppement des Plantes, CNRS IRD UP, Universite´ de Perpignan, Perpignan, France VOLUME 54 AMSTERDAM • BOSTON • HEIDELBERG • LONDON NEW YORK • OXFORD • PARIS • SAN DIEGO SAN FRANCISCO • SINGAPORE • SYDNEY • TOKYO Academic Press is an imprint of Elsevier Academic Press is an imprint of Elsevier 32 Jamestown Road, LondonNW17BY,UK 30 Corporate Drive, Suite 400, Burlington, MA 01803, USA 525 B Street, Suite 1900, San Diego, CA 92101-4495, USA Radarweg 29, PO Box 211, 1000 AE Amsterdam, The Netherlands First edition 2010 Copyright � 2010 Elsevier Ltd. All rights reserved No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means electronic, mechanical, photocopying, recording or otherwise without the prior written permission of the publisher Permissions may be sought directly from Elsevier’s Science & Technology Rights Department in Oxford, UK: phone (+44) (0) 1865 843830; fax (+44) (0) 1865 853333; email: [email protected]. Alternatively you can submit your request online by visiting the Elsevier web site at http://elsevier.com/locate/permissions, and selecting Obtaining permission to use Elsevier material Notice No responsibility is assumed by the publisher for any injury and/or damage to persons or property as a matter of products liability, negligence or otherwise, or from any use or operation of any methods, products, instructions or ideas contained in the material herein. -

What Plant Species Are Being Selected
16 Describing the world’s plants Plant genomes – progress and prospects What plant species are being selected for whole genome sequencing and why? How is the new knowledge generated from these whole genome sequences being used in both fundamental and applied biological questions? https://stateoftheworldsplants.com/2017/plant-genomics.html Plant genomes – progress and prospects 17 225 Plant Species now have whole genome sequences – and rising 18 Describing the world’s plants FIGURE 2: CURRENT USAGE OF PLANTS WHOSE THE SEQUENCE OF DNA IN PLANTS IS WHOLE GENOMES HAVE BEEN SEQUENCED A REPOSITORY OF HUGE AMOUNTS OF USEFUL INFORMATION THAT CAN BE Other PUT TO A GREAT VARIETY OF USES, 2.3% FROM ELUCIDATING THE EVOLUTIONARY RELATIONSHIPS BETWEEN DIFFERENT 37.7% PLANT SPECIES TO DETERMINING THE Model species and ORIGIN OF TROPICAL TIMBER. relatives 22.3% This year has seen huge and exciting advances in both the amounts and types of genomic data that have been Crops generated, as well as the computational approaches needed Crop wild 57.7% relatives to interpret them[e.g.1–4]. In the medical field, such advances 17.7% Human food are, for example, now starting to make personalised medicine a reality[5]. For agriculture and biodiversity, a wealth of new data promises new ways to increase food security, alleviate poverty, inform species conservation programmes 2.7% [6–10] and improve ecosystem services . In addition, these Fodder/forage advances are stimulating ever more ambitious projects Medicine in the study of evolutionary relationships across the plant Other crops 3.6% Materials Fuels 4.1% tree of life[e.g.11–14]. -

LETTER Doi:10.1038/Nature12872
LETTER doi:10.1038/nature12872 Three keys to the radiation of angiosperms into freezing environments Amy E. Zanne1,2, David C. Tank3,4, William K. Cornwell5,6, Jonathan M. Eastman3,4, Stephen A. Smith7, Richard G. FitzJohn8,9, Daniel J. McGlinn10, Brian C. O’Meara11, Angela T. Moles6, Peter B. Reich12,13, Dana L. Royer14, Douglas E. Soltis15,16,17, Peter F. Stevens18, Mark Westoby9, Ian J. Wright9, Lonnie Aarssen19, Robert I. Bertin20, Andre Calaminus15, Rafae¨l Govaerts21, Frank Hemmings6, Michelle R. Leishman9, Jacek Oleksyn12,22, Pamela S. Soltis16,17, Nathan G. Swenson23, Laura Warman6,24 & Jeremy M. Beaulieu25 Early flowering plants are thought to have been woody species to greater heights: as path lengths increase so too does resistance5. restricted to warm habitats1–3. This lineage has since radiated into Among extant strategies, the most efficient method of water delivery almost every climate, with manifold growth forms4. As angiosperms is through large-diameter water-conducting conduits (that is, vessels spread and climate changed, they evolved mechanisms to cope with and tracheids) within xylem5. episodic freezing. To explore the evolution of traits underpinning Early in angiosperm evolution they probably evolved larger conduits the ability to persist in freezing conditions, we assembled a large for water transport, especially compared with their gymnosperm cousins14. species-level database of growth habit (woody or herbaceous; 49,064 Although efficient in delivering water, these larger cells would have species), as well as leaf phenology (evergreen or deciduous), diameter impeded angiosperm colonization of regions characterized by episodic of hydraulic conduits (that is, xylem vessels and tracheids) and climate freezing14,15, as the propensity for freezing-induced embolisms (air bub- occupancies (exposure to freezing). -

Flowering Plants. Eudicots
The Families and Genera of Vascular Plants 10 Flowering Plants. Eudicots Sapindales, Cucurbitales, Myrtaceae Bearbeitet von Klaus Kubitzki 1. Auflage 2011. Buch. x, 436 S. Hardcover ISBN 978 3 642 14396 0 Format (B x L): 19,3 x 26 cm Gewicht: 1142 g Weitere Fachgebiete > Chemie, Biowissenschaften, Agrarwissenschaften > Biowissenschaften allgemein > Evolutionsbiologie Zu Inhaltsverzeichnis schnell und portofrei erhältlich bei Die Online-Fachbuchhandlung beck-shop.de ist spezialisiert auf Fachbücher, insbesondere Recht, Steuern und Wirtschaft. Im Sortiment finden Sie alle Medien (Bücher, Zeitschriften, CDs, eBooks, etc.) aller Verlage. Ergänzt wird das Programm durch Services wie Neuerscheinungsdienst oder Zusammenstellungen von Büchern zu Sonderpreisen. Der Shop führt mehr als 8 Millionen Produkte. Introduction to Sapindales K. KUBITZKI CONSPECTUS OF F AMILIES drupaceous with resinous mesocarp; seeds with oily and starchy endosperm; endotegmen lignified, 1. Herbs or low-growing shrubs 2 usually thickened; embryo curved, with fleshy – Erect shrubs or trees (some Anacardiaceae herbaceous) cotyledons.n ¼ 7–12, 14–16, 21. 81/c. 800. Pantropi- 4 cal, also temperate Anacardiaceae 2. Perennial herbs; nectary glands 5, at base of antese- 6. Fruit dehiscing with 4 or 8 one-seeded mericarps palous stamens; carpels with distinct stylodia arising from a central column; flowers isomerous, 4-merous; from base of ovarioles; ovules solitary, pendulous, testa thin; endosperm 0; trees with alternate, imparipin- epitropous; embryo sac tetrasporic, 16-celled; -

Exploring the Phylogeny of Rosids with a Five-Locus Supermatrix from Genbank
bioRxiv preprint doi: https://doi.org/10.1101/694950; this version posted July 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Exploring the phylogeny of rosids with a five-locus supermatrix from GenBank 2 Miao Sun1, 2, 3*, Ryan A. Folk1, Matthew A. Gitzendanner2, 6, Stephen A. Smith4, Charlotte 3 Germain-Aubrey1, Robert P. Guralnick1, 6, Pamela S. Soltis1, 5, 6, Douglas E. Soltis1, 2, 5, 6, 4 Zhiduan Chen3* 5 6 1. Florida Museum of Natural History, University of Florida, Gainesville, FL 32611, USA 7 2. Department of Biology, University of Florida, Gainesville, FL 32611, USA 8 3. State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, the 9 Chinese Academy of Sciences, Beijing 100093, China 10 4. Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 11 48109, USA 12 5. Genetics Institute, University of Florida, Gainesville, FL 32608, USA 13 6. Biodiversity Institute, University of Florida, Gainesville, FL 32611, USA 14 15 *Corresponding authors: 16 Miao Sun: [email protected] 17 Zhiduan Chen: [email protected] 18 1 bioRxiv preprint doi: https://doi.org/10.1101/694950; this version posted July 8, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license.