Spectral Signs of Life in Ice
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

VI FORSKER PÅ MARS Kort Om Aktiviteten I Mange Tiår Har Mars Vært Et Yndet Objekt for Forskere Verden Over
VI FORSKER PÅ MARS Kort om aktiviteten I mange tiår har Mars vært et yndet objekt for forskere verden over. Men hvorfor det? Hva er det med den røde planeten som er så interessant? Her forsøker vi å gi en oversikt over hvorfor vi er så opptatt av Mars. Hva har vi oppdaget, og hva er det vi tenker å gjøre? Det finnes en planet i solsystemet vårt som bare er bebodd av roboter -MARS- Læringsmål Elevene skal kunne - gi eksempler på dagsaktuell forskning og drøfte hvordan ny kunnskap genereres gjennom samarbeid og kritisk tilnærming til eksisterende kunnskap - utforske, forstå og lage teknologiske systemer som består av en sender og en mottaker - gjøre rede for energibevaring og energikvalitet og utforske ulike måter å omdanne, transportere og lagre energi på VI FORSKER PÅ MARS side 1 Innhold Kort om aktiviteten ................................................................................................................................ 1 Læringsmål ................................................................................................................................................ 1 Mars gjennom historien ...................................................................................................................... 3 Romkappløp mot Mars ................................................................................................................... 3 2000-tallet gir rovere i fleng ....................................................................................................... 4 Hva nå? ...................................................................................................................................................... -

Generate Viewsheds of Mastcam Images from the Curiosity Rover, Using Arcgis® and Public Datasets
TECHNICAL Coupling Mars Ground and Orbital Views: Generate REPORTS: METHODS Viewsheds of Mastcam Images From the Curiosity 10.1029/2020EA001247 Rover, Using ArcGIS® and Public Datasets Key Points: 1 2 1 3 4 • Mastcam images from the Curiosity M. Nachon , S. Borges , R. C. Ewing , F. Rivera‐Hernández , N. Stein , and rover are available online but lack a J. K. Van Beek5 public method to be placed back in the Mars orbital context 1Department of Geology and Geophysics, Texas A&M University, College Station, TX, USA, 2Department of Astronomy • This procedure allows users to and Planetary Sciences—College of Engineering, Forestry, and Natural Sciences, Northern Arizona University, Flagstaff, generate Mastcam image viewsheds: 3 4 locate in a map view the Mars AZ, USA, Department of Earth Sciences, Dartmouth College, Hanover, NH, USA, Division of Geological and Planetary 5 terrains visible in Mastcam images Sciences, California Institute of Technology, Pasadena, CA, USA, Malin Space Science Systems, San Diego, CA, USA • This procedure uses ArcGIS® and publicly available Mars datasets Abstract The Mastcam (Mast Camera) instrument onboard the NASA Curiosity rover provides an Supporting Information: exclusive view of Mars: High‐resolution color images from Mastcam allow users to study Gale crater's • Dataset S1 geologic terrains along Curiosity's path. These ground observations complement the spatially broader • Dataset S2 • Dataset S3 views of Gale crater provided by spacecrafts from orbit. However, for a given Mastcam image, it can be • Table S1 challenging to locate the corresponding terrains on the orbital view. No method for locating Mastcam images • Table S2 onto orbital images had been made publicly available. -

Mariner to Mercury, Venus and Mars
NASA Facts National Aeronautics and Space Administration Jet Propulsion Laboratory California Institute of Technology Pasadena, CA 91109 Mariner to Mercury, Venus and Mars Between 1962 and late 1973, NASA’s Jet carry a host of scientific instruments. Some of the Propulsion Laboratory designed and built 10 space- instruments, such as cameras, would need to be point- craft named Mariner to explore the inner solar system ed at the target body it was studying. Other instru- -- visiting the planets Venus, Mars and Mercury for ments were non-directional and studied phenomena the first time, and returning to Venus and Mars for such as magnetic fields and charged particles. JPL additional close observations. The final mission in the engineers proposed to make the Mariners “three-axis- series, Mariner 10, flew past Venus before going on to stabilized,” meaning that unlike other space probes encounter Mercury, after which it returned to Mercury they would not spin. for a total of three flybys. The next-to-last, Mariner Each of the Mariner projects was designed to have 9, became the first ever to orbit another planet when two spacecraft launched on separate rockets, in case it rached Mars for about a year of mapping and mea- of difficulties with the nearly untried launch vehicles. surement. Mariner 1, Mariner 3, and Mariner 8 were in fact lost The Mariners were all relatively small robotic during launch, but their backups were successful. No explorers, each launched on an Atlas rocket with Mariners were lost in later flight to their destination either an Agena or Centaur upper-stage booster, and planets or before completing their scientific missions. -

Elements of Astronomy and Cosmology Outline 1
ELEMENTS OF ASTRONOMY AND COSMOLOGY OUTLINE 1. The Solar System The Four Inner Planets The Asteroid Belt The Giant Planets The Kuiper Belt 2. The Milky Way Galaxy Neighborhood of the Solar System Exoplanets Star Terminology 3. The Early Universe Twentieth Century Progress Recent Progress 4. Observation Telescopes Ground-Based Telescopes Space-Based Telescopes Exploration of Space 1 – The Solar System The Solar System - 4.6 billion years old - Planet formation lasted 100s millions years - Four rocky planets (Mercury Venus, Earth and Mars) - Four gas giants (Jupiter, Saturn, Uranus and Neptune) Figure 2-2: Schematics of the Solar System The Solar System - Asteroid belt (meteorites) - Kuiper belt (comets) Figure 2-3: Circular orbits of the planets in the solar system The Sun - Contains mostly hydrogen and helium plasma - Sustained nuclear fusion - Temperatures ~ 15 million K - Elements up to Fe form - Is some 5 billion years old - Will last another 5 billion years Figure 2-4: Photo of the sun showing highly textured plasma, dark sunspots, bright active regions, coronal mass ejections at the surface and the sun’s atmosphere. The Sun - Dynamo effect - Magnetic storms - 11-year cycle - Solar wind (energetic protons) Figure 2-5: Close up of dark spots on the sun surface Probe Sent to Observe the Sun - Distance Sun-Earth = 1 AU - 1 AU = 150 million km - Light from the Sun takes 8 minutes to reach Earth - The solar wind takes 4 days to reach Earth Figure 5-11: Space probe used to monitor the sun Venus - Brightest planet at night - 0.7 AU from the -

Phil Liebrecht Assistant Deputy Associate Administrator and Deputy Program Manager Space Communications and Navigation NASA HQ
Phil Liebrecht Assistant Deputy Associate Administrator and Deputy Program Manager Space Communications and Navigation NASA HQ Meeting the Communications and Navigation Needs of Space missions since 1957 “Keeping the Universe Connected” Public University of Navarra, 2010 [email protected] 1 Operations and Communications Customers 2 Space Operations 101 Relation Between Space Segment, Ground System, and Data Users* Ground System Command Commands Requests Spacecraft and Payload Support • Mission Planning Telemetry • Flight Operations • Flight Dynamics • Perf. Assess., Trending & Archiving Space Data Users • Anomaly Support Segment Data Relay and Level (S/C) Mission Zero Data Processing Mission Data Data • Space-to-ground Comm. • Data Capture • Data Processing • Network Management • Data Distribution • Quick Look * Based on Wertz and Wiley; Space Mission Analysis and Design 3 Communications Theory- Basic Concepts Transmitter and Receiver must use the same language Noise causes interference a) Figures it – no errors (Identify and quantify) b) Figures it – corrects Must be loud enough ENERGY! c) Figures it – incorrectly (message in error) d) Cannot figure- recognizes E L an error T M E XP AIN L E e) Cannot figure- ? TRANSMITTER Distance weakens the sound RECEIVER (calculate the loss) CHANNEL Has to correctly interpret the message - the most difficult job The fundamental problem of communications is that of reproducing at one point either exactly or approximately a message selected at another point. 4 Functional - End to End Process -
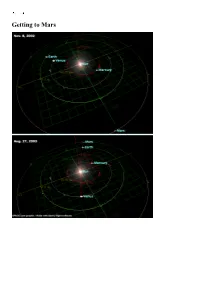
Getting to Mars How Close Is Mars?
Getting to Mars How close is Mars? Exploring Mars 1960-2004 Of 42 probes launched: 9 crashed on launch or failed to leave Earth orbit 4 failed en route to Mars 4 failed to stop at Mars 1 failed on entering Mars orbit 1 orbiter crashed on Mars 6 landers crashed on Mars 3 flyby missions succeeded 9 orbiters succeeded 4 landers succeeded 1 lander en route Score so far: Earthlings 16, Martians 25, 1 in play Mars Express Mars Exploration Rover Mars Exploration Rover Mars Exploration Rover 1: Meridiani (Opportunity) 2: Gusev (Spirit) 3: Isidis (Beagle-2) 4: Mars Polar Lander Launch Window 21: Jun-Jul 2003 Mars Express 2003 Jun 2 In Mars orbit Dec 25 Beagle 2 Lander 2003 Jun 2 Crashed at Isidis Dec 25 Spirit/ Rover A 2003 Jun 10 Landed at Gusev Jan 4 Opportunity/ Rover B 2003 Jul 8 Heading to Meridiani on Sunday Launch Window 1: Oct 1960 1M No. 1 1960 Oct 10 Rocket crashed in Siberia 1M No. 2 1960 Oct 14 Rocket crashed in Kazakhstan Launch Window 2: October-November 1962 2MV-4 No. 1 1962 Oct 24 Rocket blew up in parking orbit during Cuban Missile Crisis 2MV-4 No. 2 "Mars-1" 1962 Nov 1 Lost attitude control - Missed Mars by 200000 km 2MV-3 No. 1 1962 Nov 4 Rocket failed to restart in parking orbit The Mars-1 probe Launch Window 3: November 1964 Mariner 3 1964 Nov 5 Failed after launch, nose cone failed to separate Mariner 4 1964 Nov 28 SUCCESS, flyby in Jul 1965 3MV-4 No. -

MR PRISM – Spectral Analysis Tool for the CRISM
MR PRISM – Spectral Analysis Tool for the CRISM Adrian J. Brown*a, and Michael Storrie-Lombardib a SETI Institute, 515 N. Whisman Rd, Mountain View, CA 94043 b Kinohi Institute, Pasadena, CA 91101 ABSTRACT We describe a computer application designed to analyze hyperspectral data collected by the Compact Infrared Spectrometer for Mars (CRISM). The application links the spectral, imaging and mapping perspectives on the eventual CRISM dataset by presenting the user with three different ways to analyze the data. One of the goals when developing this instrument is to build in the latest algorithms for detection of spectrally compelling targets on the surface of the Red Planet, so they may be available to the Planetary Science community without cost and with a minimal learning barrier to cross. This will allow the Astrobiology community to look for targets of interest such as hydrothermal minerals, sulfate minerals and hydrous minerals and be able to map the extent of these minerals using the most up-to-date and effective algorithms. The application is programmed in Java and will be made available for Windows, Mac and Linux platforms. Users will be able to embed Groovy scripts into the program in order to extend its functionality. The first collection of CRISM data will occur in September of 2006 and this data will be made publicly available six months later via the Planetary Datasystem (PDS). Potential users in the community should therefore look forward to a release date mid-2007. Although exploration of the CRISM data set is the motivating force for developing these software tools, the ease of writing additional Groovy scripts to access other data sets makes the tools useful for mineral exploration, crop management, and characterization of extreme environments here on Earth or other terrestrial planets. -

Composition of Mars, Michelle Wenz
The Composition of Mars Michelle Wenz Curiosity Image NASA Importance of minerals . Role in transport and storage of volatiles . Ex. Water (adsorbed or structurally bound) . Control climatic behavior . Past conditions of mars . specific pressure and temperature formation conditions . Constrains formation and habitability Curiosity Rover at Mount Sharp drilling site, NASA image Missions to Mars . 44 missions to Mars (all not successful) . 21 NASA . 18 Russia . 1 ESA . 1 India . 1 Japan . 1 joint China/Russia . 1 joint ESA/Russia . First successful mission was Mariner 4 in 1964 Credit: Jason Davis / astrosaur.us, http://utprosim.com/?p=808 First Successful Mission: Mariner 4 . First image of Mars . Took 21 images . No evidence of canals . Not much can be said about composition Mariner 4, NASA image Mariner 4 first image of Mars, NASA image Viking Lander . First lander on Mars . Multispectral measurements Viking Planning, NASA image Viking Anniversary Image, NASA image Viking Lander . Measured dust particles . Believed to be global representation . Computer generated mixtures of minerals . quartz, feldspar, pyroxenes, hematite, ilmenite Toulmin III et al., 1977 Hubble Space Telescope . Better resolution than Mariner 6 and 7 . Viking limited to three bands between 450 and 590 nm . UV- near IR . Optimized for iron bearing minerals and silicates Hubble Space Telescope NASA/ESA Image featured in Astronomy Magazine Hubble Spectroscopy Results . 1994-1995 . Ferric oxide absorption band 860 nm . hematite . Pyroxene 953 nm absorption band . Looked for olivine contributions . 1042 nm band . No significant olivine contributions Hubble Space Telescope 1995, NASA Composition by Hubble . Measure of the strength of the absorption band . Ratio vs. -

Mars Science Laboratory XXX XX XXXX
Exploring Mars with CuriosityChemCam and its Laser-Induced Remote Sensing for ChemistryLaser and Micro-Imaging (meeting) Mg Si Roger C Wiens Al Ba ChemCam PI Frontiers in Science Lectures Los Alamos – Albuquerque – Santa Fe – Taos (location) 3/16/2004 1 NASA (date)May, 2013 LA-UR-13-23209 Jean-Luc Lacour, CEA Mars Science Laboratory Goals • Assess Mars’ biological potential by • Searching for organic carbon compounds, • Looking for the chemical building blocks of life, • Identify biologically relevant clues. • Characterize the geology of the landing region • Investigate Mars’ past habitability (including the role of water) • Characterize the human hazards on Mars Spirit, Opportunity 2003 Sojourner 1997 MSL 2011 2 NASA/JPL-Caltech Is Mars Like Earth? • Starting 150 years ago, some astronomers thought they could see canals on Mars Schiaparelli 3 Mariners 4, 6, 7 Flyby Missions Mariner 4, 1965: Mars looks cratered and barren, like the Moon 4 NASA Mariner 9 Orbiter, 1971: Evidence of Water 5 NASA Viking Landers, 1976, Test for Life 6 NASA Biota Could Have Traveled From Earth to Mars • NASA has > 100 meteorites from Mars that fell to Earth • Mars probably has a number of meteorites from Earth – Bacteria from Earth could have traveled in these rocks NASA/JSC/Smithsonian 7 Fossilization How to Look for Life on Mars • Concentration: where does water deposit material? – Lakebeds and especially river deltas – Coal- or oil-bearing stata on Earth • Preservation : mineralization = fossils – Sedimentary rocks, especially clay- bearing Coal R. Wiens, -

Aerospace Micro-Lesson
AIAA AEROSPACE M ICRO-LESSON Easily digestible Aerospace Principles revealed for K-12 Students and Educators. These lessons will be sent on a bi-weekly basis and allow grade-level focused learning. - AIAA STEM K-12 Committee. THE MARINER PROJECT Started less than five years after the first launch of rockets into orbit, the Project Mariner was the United States’ first attempt to send a spacecraft to another planet. Between 1962 and 1975, Project Mariner involved the launch of ten unmanned spacecraft, seven of them successfully, to the other inner planets of the Solar System. You can find an overview of the project at NASA’s Science web site. GRADES K-2 Astronomy has always been about looking at things from a distance. For centuries and longer, stars and planets were simply points of light in the sky, most of them stationary and a few of them moving. (In fact, the word “planet” is derived from the Greek word “planetes,” which means “wanderer.”) The invention of the telescope changed the view of planets from wandering points of light to bodies that move around the Sun, but even the best telescopes could not distinguish features smaller than a mile across on the moon and several tens of miles across on even the closest planets. The space program changed this. By sending spacecraft to visit the moon and the planets, scientists were able to view these bodies from comparatively close up and see them in much greater detail. This revolutionized solar system astronomy and gave birth to a new science, “planetary science.” Planetary science joins the fields of astronomy, geology, meteorology, and others together into a unified study of the planets. -

The Search and Prospects for Life on Mars Overview
AST 309 part 2: Extraterrestrial Life The Search and Prospects for Life on Mars Overview: • Life on Mars: a historical view • Present-day Mars • The Exploration of Mars • The Viking Mission • AHL 84001 • Mars Exploration Rovers • Evidence for water • Methane on Mars! • The future Our perception of Mars through history: • named after the roman god of war (probably due to its color) • (mainly) used by Johannes Kepler to derive laws of planetary motion • early observations with telescopes showed polar caps, dark areas and moons => very Earth-like? • Percival Lowell: canals on Mars? Intelligent life? Lowell in the 1890s Our perception of Mars through history: H.G. Wells: “The War of the Worlds” (1898) Orson Wells’ radio broadcast in 1938 Modern exploration of Mars: Mariner 4 Early missions: 1964 Mariner 4 first flyby 1971 Mariner 9 orbits Mars Mars 3 & 4 land (but stopped working) 1976 2 Viking orbiter + Landers 1988 Phobos 1 & 2 failed 1992 Mars Observer failed 1997 Mars Global Surveyor & Mars Pathfinder begin modern era Olympus Mons Mars basic facts: Distance: 1.5 AU Period: 1.87 years Radius: 0.53 R_Earth Mass: 0.11 M_Earth Density: 4.0 g/cm3 Satellites: Phobos and Deimos Structure: Dense Core (~1700 km), rocky mantle, thin crust Temperature: -87 to -5 C Magnetic Field: Weak and variable (some parts strong) Atmosphere: 95% CO2, 3% Nitrogen, argon, traces of oxygen Mars atmosphere: Very thin! ~0.7% of Earth’s pressure Pressure can change significantly because it gets so cold that CO2 freezes out on polar caps Viking atmospheric measurements 95.3% carbon dioxide 2.7% nitrogen 1.6% argon 0.13% oxygen Composition 0.07% carbon monoxide 0.03% water vapor trace neon, krypton, xenon, ozone, methane 1-9 millibars, depending on Surface pressure altitude; average 7 mb Viking on Mars: Viking 1 Lander touched down at Chryse Planitia (22.48° N, 49.97° W planetographic, 1.5 km below the datum (6.1 mbar) elevation). -

ISPRS Working Group IV/8 Planetary Mapping and Spatial Databases Meeting 24 – 25 September 2015, Berlin, Germany
ISPRS Working Group IV/8 Planetary Mapping and Spatial Databases Meeting 24 – 25 September 2015, Berlin, Germany MAPPING OF SWISS CHEESE TERRAIN USING HRSC AND CRISM DATA Jacqueline Campbell (1), (2) Panagiotis Sidiropoulos (2) and Jan-Peter Muller (2) (1) School of Environment and Technology, University of Brighton, East Sussex, BN2 4AT, [email protected] and (2) Imaging Group, Mullard Space Science Laboratory, University College London, Holmbury St Mary, Surrey, RH56NT, [email protected], [email protected] ABSTRACT: Since 1965 and NASA’s Mariner 4, Mars has been frequently examined via high resolution orbital imagery, and with spectrometers since Mariner 6 in 1969 (NASA, 2015). The dynamic polar regions of Mars have been systematically studied in detail in more recent years by the European Space Agency’s (ESA) 2003 Mars Express and NASA’s Mars Reconnaissance Orbiter (MRO) launched in 2005; in particular, Mars’ south polar cap consists of a permanent 400km diameter layer of solid CO2 and water ice (Vita-Finzi, 2005). Swiss Cheese Terrain is a unique surface feature found only in the Martian south polar ice. Its characteristic appearance (consisting of flat floored, circular depressions) is considered to be caused by seasonal differences in the sublimation rates of water and CO2 ice (Tokar et al. 2003). A number of researchers have examined the properties of Swiss Cheese Terrain; Brown et al. (2014) examined H2O ice deposition on the Martian south pole, using measurements from the OMEGA infrared spectrometer on board Mars Express and the Compact Reconnaissance Imaging Spectrometer for Mars (CRISM) on MRO to measure seasonal changes in water ice signatures around Swiss Cheese depressions.