Molecular Properties of Membrane-Bound FAD-Containing D-Sorbitol Dehydrogenase from Thermotolerant Gluconobacter Frateurii Isolated from Thailand
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glycerol Dehydrogenase from Gluconobacter Industrius
Agric. Biol Chem., 49 (4), 1001 -1010, 1985 1001 Solubilization, Purification and Properties of Membrane-bound Glycerol Dehydrogenase from Gluconobacter industrius Minoru Ameyama,Emiko Shinagawa, Kazunobu Matsushita and Osao Adachi Laboratory of Applied Microbiology, Department of Agricultural Chemistry, Faculty of Agriculture, Yamaguchi University, Yamaguchi 753, Japan Received July 30, 1984 Membrane-bound glycerol dehydrogenase was solubilized and purified about 100-fold from the membraneof Gluconobacter industrius IFO 3260 grown on a glycerol-glutamate medium. Solubilization of the enzyme was successfully achieved by use of 0.5% dimethyldodecylamineoxide in 0.05 m Tris-HCl, pH 8.0. Alcohol dehydrogenase and D-glucose dehydrogenase, which were abundantly formed in the same bacterial membrane, were eliminated on solubilization. Glycerol dehydrogenase was further purified through fractionation with polyethylene glycol 6000. The enzymeshowed a broad substrate specificity and various kinds of polyhydroxyl alcohols, in addition to glycerol, were rapidly oxidized in the presence of 2,6-dichlorophenolindophenoi and phenazine methosulfate as the electron acceptor but NADand NADPwere inert. The enzyme was proved to be a quinoprotein in which pyrroloquinoline quinone functioned as the prosthetic group. The first report on microbial oxidation of localization of the oxidase system in cells of G. glycerol to dihydroxyacetone was by Bertrand liquefaciens and found that the oxidation of with a strain capable of L-sorbose fermen- glycerol and raeso-erythritol -

Clostridium Difficile Exploits a Host Metabolite Produced During Toxin-Mediated Infection
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.14.426744; this version posted January 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Clostridium difficile exploits a host metabolite produced during toxin-mediated infection 2 3 Kali M. Pruss and Justin L. Sonnenburg* 4 5 Department of Microbiology & Immunology, Stanford University School of Medicine, Stanford 6 CA, USA 94305 7 *Corresponding author address: [email protected] 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 bioRxiv preprint doi: https://doi.org/10.1101/2021.01.14.426744; this version posted January 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 23 Several enteric pathogens can gain specific metabolic advantages over other members of 24 the microbiota by inducing host pathology and inflammation. The pathogen Clostridium 25 difficile (Cd) is responsible for a toxin-mediated colitis that causes 15,000 deaths in the U.S. 26 yearly1, yet the molecular mechanisms by which Cd benefits from toxin-induced colitis 27 remain understudied. Up to 21% of healthy adults are asymptomatic carriers of toxigenic 28 Cd2, indicating that Cd can persist as part of a healthy microbiota; antibiotic-induced 29 perturbation of the gut ecosystem is associated with transition to toxin-mediated disease. -

Ular Basis of KAISER, S
Heredity 74 (1995) 267—273 Received6May 1994 OThe Genetical Society of Great Britain 1981. The molecular basis of KAISER, S. 1935. The factorsSOMMER, controlling H. 1991. Genetic shape control ofand flower sizedevelopment in Genetic differentiation among populations of by homeotic genes in Antirrhinum majus. Science, 250, Myopus schisticolor (the wood lemming) — LANDE, R. 1981. The minimumSHORE, number J. S. AND BARRETF, of genes S. C. H. 1990. contributing Quantitative genetics of floral characters in homostylous Turnera ulmifolia var, isozyme variation angustifolia Willd. (Turneraceae). Heredity, 64, LANDE, R, 1983. The responseSINNO1F, to e. w.selection 1935. Evidence onfor the major existence of and genes VADIM B. FEDOROVif, RICKARD FREDRIKSSON1: & KARLFREDGA* SINNO'rr, E. w. 1936. A developmental analysis of inherited t/nstitute of Plant and Animal Ecology, Russian Academy of Sciences, 8 March Street 202, Jekaterinburg 620 008, tion on correlated characters.shape Evolution, differences in Cucurbit37, 1210—1226. fruits. Am. Nat., 70, Russia and Department of Genetics, Uppsa/a University, Box 7003, S-75007Uppsala, Sweden LORD, C. M. AND HILL, .i. i'. 1987. Evidence for heterochrony in Toinvestigate genetic differentiation among populations of the wood lemming Myopus schistico!or (eds) Development as an Evolutiona,ySMITH, H. H. 1950. DevelopmentalProcess, restrictions pp. 47—70. on recombina- (Muridae, Rodentia) isozyme variation in 10 populations from three regions, Fennoscandia, Western and Eastern Siberia, was examined. From 20 loci examined, the two most polymorphic MACNAIR, M. R. AND CUMBES,TAKHTAJAN, o. .i. 1989. A. 1976. The Neoteny genetic and the architectureorigin of flowering ones, Idh-1 and Pgi-1, were used in the complete survey. -

High-Yield Anaerobic Succinate Production by Strategically
Meng et al. Microb Cell Fact (2016) 15:141 DOI 10.1186/s12934-016-0536-1 Microbial Cell Factories RESEARCH Open Access High‑yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli Jiao Meng1,2,3†, Baiyun Wang1,2,3†, Dingyu Liu1,2,3, Tao Chen1,2,3,4, Zhiwen Wang1,2,3* and Xueming Zhao1,2,3 Abstract Background: Succinate has been identified by the U.S. Department of Energy as one of the top 12 building block chemicals, which can be used as a specialty chemical in the agricultural, food, and pharmaceutical industries. Escheri- chia coli are now one of the most important succinate producing candidates. However, the stoichiometric maximum succinate yield under anaerobic conditions through the reductive branch of the TCA cycle is restricted by NADH sup- ply in E. coli. Results: In the present work, we report a rational approach to increase succinate yield by regulating NADH supply via pentose phosphate (PP) pathway and enhancing flux towards succinate. The deregulated genes zwf243 (encod- ing glucose-6-phosphate dehydrogenase) and gnd361 (encoding 6-phosphogluconate dehydrogenase) involved in NADPH generation from Corynebacterium glutamicum were firstly introduced into E. coli for succinate production. Co-expression of beneficial mutated dehydrogenases, which removed feedback inhibition in the oxidative part of the PP pathway, increased succinate yield from 1.01 to 1.16 mol/mol glucose. Three critical genes, pgl (encoding 6-phos- phogluconolactonase), tktA (encoding transketolase) and talB (encoding transaldolase) were then overexpressed to redirect more carbon flux towards PP pathway and further improved succinate yield to 1.21 mol/mol glucose. -

Global Profiling of Metabolic Adaptation to Hypoxic Stress in Human Glioblastoma Cells
Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. Kucharzewska, Paulina; Christianson, Helena; Belting, Mattias Published in: PLoS ONE DOI: 10.1371/journal.pone.0116740 2015 Link to publication Citation for published version (APA): Kucharzewska, P., Christianson, H., & Belting, M. (2015). Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLoS ONE, 10(1), [e0116740]. https://doi.org/10.1371/journal.pone.0116740 Total number of authors: 3 General rights Unless other specific re-use rights are stated the following general rights apply: Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal Read more about Creative commons licenses: https://creativecommons.org/licenses/ Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. LUND UNIVERSITY PO Box 117 221 00 Lund +46 46-222 00 00 Download date: 07. Oct. 2021 RESEARCH ARTICLE Global Profiling of Metabolic Adaptation to Hypoxic Stress in Human Glioblastoma Cells Paulina Kucharzewska1, Helena C. -

The Diversity of Microbial Aldo/Keto Reductases from Escherichia Coli K12
Lapthorn, A. J., Zhu, X., and Ellis, E. M. (2013) The diversity of microbial aldo/keto reductases from Escherichia coli K12. Chemico-Biological Interactions, 202(1-3), pp. 168-177. (doi:10.1016/j.cbi.2012.10.008) There may be differences between this version and the published version. You are advised to consult the publisher’s version if you wish to cite from it. http://eprints.gla.ac.uk/124204/ Deposited on: 07 October 2016 Enlighten – Research publications by members of the University of Glasgow http://eprints.gla.ac.uk The diversity of microbial aldo/keto reductases from Escherichia coli K12 Adrian J. Lapthorn 1, Xiaofeng Zhu 2,3 and Elizabeth M. Ellis 3 1 School of Chemistry, Joseph Black Building, University of Glasgow, Glasgow G12 8QQ 2 College of Life Science and State Key Laboratory of Biotherapy and Cancer Centre, Sichuan University, Chengdu, China 3 Strathclyde Institute of Pharmacy and Biomedical Sciences, 161 Cathedral Street, Glasgow, G4 0RE Corresponding author: Adrian J. Lapthorn School of Chemistry, Joseph Black Building, University of Glasgow, Glasgow G12 8QQ Tel: +44 141-330 5940 Fax: +44 141-330 4888 E-mail: [email protected] Key Words: Aldo-Keto reductases, methylglyoxal reductase, 2,5-diketo-D-gluconate reductase, tyrosine auxotrophy suppressor protein, L-glyceraldehyde 3-phosphate reductase, AKR quaternary structure. Abstract The genome of Escherichia coli K12 contains 9 open reading frames encoding aldo/keto reductases (AKR) that are differentially regulated and sequence diverse. A significant amount of data is available for the E. coli AKRs through the availability of gene knockouts and gene expression studies, which adds to the biochemical and kinetic data. -

Engineering of Glycerol Utilization in Gluconobacter Oxydans 621H For
Yan et al. Microb Cell Fact (2018) 17:158 https://doi.org/10.1186/s12934-018-1001-0 Microbial Cell Factories RESEARCH Open Access Engineering of glycerol utilization in Gluconobacter oxydans 621H for biocatalyst preparation in a low‑cost way Jinxin Yan1, Jing Xu1,3, Menghao Cao1, Zhong Li1, Chengpeng Xu1, Xinyu Wang1, Chunyu Yang1, Ping Xu2, Chao Gao1 and Cuiqing Ma1* Abstract Background: Whole cells of Gluconobacter oxydans are widely used in various biocatalytic processes. Sorbitol at high concentrations is commonly used in complex media to prepare biocatalysts. Exploiting an alternative process for preparation of biocatalysts with low cost substrates is of importance for industrial applications. Results: G. oxydans 621H was confrmed to have the ability to grow in mineral salts medium with glycerol, an inevitable waste generated from industry of biofuels, as the sole carbon source. Based on the glycerol utilization mechanism elucidated in this study, the major polyol dehydrogenase (GOX0854) and the membrane-bound alcohol dehydrogenase (GOX1068) can competitively utilize glycerol but play no obvious roles in the biocatalyst prepara- tion. Thus, the genes related to these two enzymes were deleted. Whole cells of G. oxydans ∆GOX1068∆GOX0854 can be prepared from glycerol with a 2.4-fold higher biomass yield than that of G. oxydans 621H. Using whole cells of G. 1 1 oxydans ∆GOX1068∆GOX0854 as the biocatalyst, 61.6 g L− xylonate was produced from 58.4 g L− xylose at a yield of 1 1.05 g g− . Conclusion: This process is an example of efcient preparation of whole cells of G. oxydans with reduced cost. -

Characterization of Glycerol Dehydrogenase from Thermoanaerobacterium Thermosaccharolyticum DSM 571 and GGG Motif Identification
J. Microbiol. Biotechnol. (2016), 26(6), 1077–1086 http://dx.doi.org/10.4014/jmb.1512.12051 Research Article Review jmb Characterization of Glycerol Dehydrogenase from Thermoanaerobacterium thermosaccharolyticum DSM 571 and GGG Motif Identification Liangliang Wang1,2, Jiajun Wang1,2, Hao Shi1,2, Huaxiang Gu1,2, Yu Zhang1,2, Xun Li1,2, and Fei Wang1,2* 1College of Chemical Engineering, Nanjing Forestry University, Nanjing 210037, P.R. China 2Jiangsu Key Laboratory of Biomass-Based Green Fuels and Chemicals, Nanjing 210037, P.R. China Received: December 17, 2015 Revised: March 5, 2016 Glycerol dehydrogenases (GlyDHs) are essential for glycerol metabolism in vivo, catalyzing Accepted: March 9, 2016 its reversible reduction to 1,3-dihydroxypropranone (DHA). The gldA gene encoding a putative GlyDH was cloned from Thermoanaerobacterium thermosaccharolyticum DSM 571 (TtGlyDH) and expressed in Escherichia coli. The presence of Mn2+ enhanced its enzymatic 171 254 271 First published online activity by 79.5%. Three highly conserved residues (Asp , His , and His ) in TtGlyDH were March 14, 2016 associated with metal ion binding. Based on an investigation of glycerol oxidation and DHA *Corresponding author reduction, TtGlyDH showed maximum activity towards glycerol at 60°C and pH 8.0 and Phone: +86-25-85427649; towards DHA at 60°C and pH 6.0. DHA reduction was the dominant reaction, with a lower Fax: +86-25-85427649; K of 1.08 ± 0.13 mM and V of 0.0053 ± 0.0001 mM/s, compared with glycerol oxidation, E-mail: [email protected] m(DHA) max with a Km(glycerol) of 30.29 ± 3.42 mM and Vmax of 0.042 ± 0.002 mM/s. -

Polyol Pathway Links Glucose Metabolism to the Aggressiveness
Published OnlineFirst January 17, 2018; DOI: 10.1158/0008-5472.CAN-17-2834 Cancer Metabolism and Chemical Biology Research Polyol Pathway Links Glucose Metabolism to the Aggressiveness of Cancer Cells Annemarie Schwab1, Aarif Siddiqui1, Maria Eleni Vazakidou1, Francesca Napoli1, Martin Bottcher€ 2, Bianca Menchicchi3, Umar Raza4, Ozge€ Saatci4, Angela M. Krebs5, Fulvia Ferrazzi6, Ida Rapa7, Katja Dettmer-Wilde8, Maximilian J. Waldner3, Arif B. Ekici6, Suhail Ahmed Kabeer Rasheed9, Dimitrios Mougiakakos2, Peter J. Oefner8, Ozgur Sahin4, Marco Volante7, Florian R. Greten10, Thomas Brabletz5, and Paolo Ceppi1 Abstract Cancer cells alter their metabolism to support their malig- sequencing confirmed a profound alteration of EMT in PP- nant properties. In this study, we report that the glucose- deficient cells, revealing a strong repression of TGFb signature transforming polyol pathway (PP) gene aldo-keto-reductase- genes. Excess glucose was found to promote EMT through 1-member-B1 (AKR1B1) strongly correlates with epithelial-to- autocrine TGFb stimulation, while PP-deficient cells were mesenchymal transition (EMT). This association was con- refractory to glucose-induced EMT. These data show that PP firmed in samples from lung cancer patients and from an represents a molecular link between glucose metabolism, can- EMT-driven colon cancer mouse model with p53 deletion. In cer differentiation, and aggressiveness, and may serve as a novel vitro, mesenchymal-like cancer cells showed increased AKR1B1 therapeutic target. levels, and AKR1B1 knockdown was sufficient to revert EMT. An Significance: A glucose-transforming pathway in TGFb-driven equivalent level of EMT suppression was measured by targeting epithelial-to-mesenchymal transition provides novel mecha- the downstream enzyme sorbitol-dehydrogenase (SORD), fur- nistic insights into the metabolic control of cancer differenti- ther pointing at the involvement of the PP. -

Sorbitol Dehydrogenase (SDH) Polyol Dehydrogenase from Sheep Liver L-Iditol: NAD 5´-Oxidoreductase, EC 1.1.1.14
For life science research only. Not for use in diagnostic procedures. Sorbitol Dehydrogenase (SDH) Polyol dehydrogenase from sheep liver L-Iditol: NAD 5´-oxidoreductase, EC 1.1.1.14 Cat. No. 10 109 339 001 10 mg (60 mg lyo.) y Version 06 Content version: June 2019 Store at +2 to +8°C Product overview • In the colorimetric assay6 of sorbitol and xylitol, high concentrations of reducing substances Ն Formulation Lyophilizate (12 mg contain 2 mg enzyme protein and ( 5 g/assay) such as ascorbic acid (in fruit juice) 10 mg maltose; 60 mg contain 10 mg enzyme protein or SO2 (in jam) interfere. A procedure for removing and 50 mg maltose). these reducing substances (with H2O2 and alkali) is given in reference6. Contaminants ADH < 0.01%, GIDH < 0.02%, glucose dehydrogenase < 0.02%, Analysis Information LDH < 0.05%, MDH < 0.05% Quality Control Mr 115,000 SDH Substrate Sorbitol dehydrogenase (SDH) will oxidize D-sorbitol to D-fructose + NADH + H+ D-sorbitol + specificity, relative fructose (Km = 0.7 mM; relative rate = 1.00). The + NAD rates and Km enzyme will also oxidize many other polyols, including L-iditiol to L-sorbose (rate = 0.96), xylitol to D-xylulose (rate = 0.85), ribitol to D-ribulose (rate = 0.49) and Unit definition One unit (U) sorbitol dehydrogenase will reduce allitol to allulose (rate = 0.45). SDH also catalyzes the 1 mol of D-fructose in 1 min at 25° C and pH 7.6 reverse (reduction) reactions of each of the above. The [triethanolamine buffer; 150 mM fructose (non- Km for fructose is 250-300 mM. -

Transcriptional Regulation Mechanisms Involved in Azole Resistance in Candida Species: Focusing on the Transcription Factors Rpn4 and Mrr1
Transcriptional regulation mechanisms involved in azole resistance in Candida species: focusing on the transcription factors Rpn4 and Mrr1 Raquel da Silva Califórnia Thesis to obtain the Master of Science Degree in Biotechnology Supervisor: Prof. Dr. Miguel Nobre Parreira Cacho Teixeira Examination Committee Chairperson: Prof. Dr. Ana Cristina Anjinho Madeira Viegas Supervisor: Prof. Dr. Miguel Nobre Parreira Cacho Teixeira Member of the Committee: Dr. Catarina Isabel Ribeiro Pimentel October 2018 ii Acknowledgements For me the development of this thesis was very challenging and involved a very extensive work, whose purpose would not have been reached without the help of some people I will mention below. First of all, I would like to thank my supervisor Professor Miguel Teixeira for the opportunity given by accepting me in his team and in this project. His tremendous support, guidance and motivation, always available to help, were crucial for the success of this work. I would like to thank Professor Isabel Sá-Correia for giving me the chance to join the Biological Sciences Research Group to develop my master thesis work. The achievement of this thesis required an indispensable help from several parts, which deserve my recognition. For the collaboration in the transcriptomic analysis herein accomplished, I thank Professor Geraldine Butler and her team, from University College of Dublin. For the supply of Candida glabrata mutants used in this work, I have to thank Professor Hiroji Chibana, from University of Chiba, Japan. For the study developed in HPLC analysis of ergosterol levels, I thank also Professor Nuno Mira for his availability and assistance. My gratitude should also be expressed towards my colleague, Pedro Pais, for the great help he has given me throughout this period, always available to help and explain anything. -
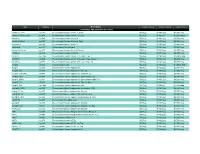
Prospec (Product
Name Catalog # Description Customer Price-A Customer Price-B Customer Price-C CYTOKINES AND GROWTH FACTORS Activin A, Active cyt-145 Recombinant Human Activin-A, Active $50/2µg $130/10µg $3,500/1mg Activin A, Plant-Active cyt-414 Recombinant Human Activin-A Active $50/1µg $130/5µg $1,500/100µg Activin A cyt-569 Recombinant Human Activin-A $50/2µg $130/10µg $2,700/1mg Activin A, Plant cyt-052 Recombinant Human Activin-A, Plant $50/2µg $130/10µg $4,800/1mg mActivin A cyt-146 Recombinant Mouse Activin-A $50/2µg $130/10µg $3,500/1mg rActivin A cyt-147 Recombinant Rat Activin-A $50/2µg $130/10µg $3,500/1mg Activin B, Active cyt-057 Recombinant Human Activin-B Active $50/2µg $130/10µg $5,200/1mg Activin B cyt-058 Recombinant Human Activin-B $50/2µg $130/10µg $4,800/1mg ACVR1 cyt-1140 Recombinant Human Activin A Receptor Type 1 $50/2µg $130/10µg $1,000/0.1mg ACVRL1 cyt-920 Recombinant Human Activin A Receptor Type II-Like 1 $50/2µg $130/10µg $5,200/1mg ACVR2A cyt-976 Recombinant Human Activin A Receptor Type 2A $50/2µg $130/10µg $5,200/1mg Acrp30 cyt-024 Human Adiponectin $50/2µg $130/10µg $1,000/0.1mg Acrp30 cyt-280 Recombinant Human Adiponectin $50/5µg $130/25µg $2,700/1mg Acrp30, His cyt-433 Recombinant Human Adiponectin, His tag $50/10µg $130/50µg $1,900/1mg Acrp30 (108-244) cyt-073 Recombinant Human Adiponectin (108-244 a.a.) $50/5µg $130/25µg $2,700/1mg Acrp30, HEK cyt-434 Recombinant Human Adiponectin glycosilated, HEK $50/2µg $130/10µg $4,680/1mg Acrp30, HMW cyt-764 Recombinant Human Adiponectin glycosilated, HMW Rich $50/2µg $130/10µg