Mouse Mthfd2l Knockout Project (CRISPR/Cas9)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Folate Cycle Enzyme MTHFD1L Confers Metabolic Advantages in Hepatocellular Carcinoma
RESEARCH ARTICLE The Journal of Clinical Investigation Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma Derek Lee,1 Iris Ming-Jing Xu,1 David Kung-Chun Chiu,1 Robin Kit-Ho Lai,1 Aki Pui-Wah Tse,1 Lynna Lan Li,1 Cheuk-Ting Law,1 Felice Ho-Ching Tsang,1 Larry Lai Wei,1 Cerise Yuen-Ki Chan,1 Chun-Ming Wong,1,2 Irene Oi-Lin Ng,1,2 and Carmen Chak-Lui Wong1,2 1Department of Pathology and 2State Key Laboratory for Liver Research, The University of Hong Kong, Hong Kong, China. Cancer cells preferentially utilize glucose and glutamine, which provide macromolecules and antioxidants that sustain rapid cell division. Metabolic reprogramming in cancer drives an increased glycolytic rate that supports maximal production of these nutrients. The folate cycle, through transfer of a carbon unit between tetrahydrofolate and its derivatives in the cytoplasmic and mitochondrial compartments, produces other metabolites that are essential for cell growth, including nucleotides, methionine, and the antioxidant NADPH. Here, using hepatocellular carcinoma (HCC) as a cancer model, we have observed a reduction in growth rate upon withdrawal of folate. We found that an enzyme in the folate cycle, methylenetetrahydrofolate dehydrogenase 1–like (MTHFD1L), plays an essential role in support of cancer growth. We determined that MTHFD1L is transcriptionally activated by NRF2, a master regulator of redox homeostasis. Our observations further suggest that MTHFD1L contributes to the production and accumulation of NADPH to levels that are sufficient to combat oxidative stress in cancer cells. The elevation of oxidative stress through MTHFD1L knockdown or the use of methotrexate, an antifolate drug, sensitizes cancer cells to sorafenib, a targeted therapy for HCC. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -
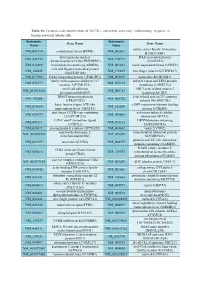
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Molecular Signatures Differentiate Immune States in Type 1 Diabetes Families
Page 1 of 65 Diabetes Molecular signatures differentiate immune states in Type 1 diabetes families Yi-Guang Chen1, Susanne M. Cabrera1, Shuang Jia1, Mary L. Kaldunski1, Joanna Kramer1, Sami Cheong2, Rhonda Geoffrey1, Mark F. Roethle1, Jeffrey E. Woodliff3, Carla J. Greenbaum4, Xujing Wang5, and Martin J. Hessner1 1The Max McGee National Research Center for Juvenile Diabetes, Children's Research Institute of Children's Hospital of Wisconsin, and Department of Pediatrics at the Medical College of Wisconsin Milwaukee, WI 53226, USA. 2The Department of Mathematical Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI 53211, USA. 3Flow Cytometry & Cell Separation Facility, Bindley Bioscience Center, Purdue University, West Lafayette, IN 47907, USA. 4Diabetes Research Program, Benaroya Research Institute, Seattle, WA, 98101, USA. 5Systems Biology Center, the National Heart, Lung, and Blood Institute, the National Institutes of Health, Bethesda, MD 20824, USA. Corresponding author: Martin J. Hessner, Ph.D., The Department of Pediatrics, The Medical College of Wisconsin, Milwaukee, WI 53226, USA Tel: 011-1-414-955-4496; Fax: 011-1-414-955-6663; E-mail: [email protected]. Running title: Innate Inflammation in T1D Families Word count: 3999 Number of Tables: 1 Number of Figures: 7 1 For Peer Review Only Diabetes Publish Ahead of Print, published online April 23, 2014 Diabetes Page 2 of 65 ABSTRACT Mechanisms associated with Type 1 diabetes (T1D) development remain incompletely defined. Employing a sensitive array-based bioassay where patient plasma is used to induce transcriptional responses in healthy leukocytes, we previously reported disease-specific, partially IL-1 dependent, signatures associated with pre and recent onset (RO) T1D relative to unrelated healthy controls (uHC). -

Differentially Expressed Genes Yuki
supplementary Table 1. The differentially expressed genes in F3-treated THP-1 cells after 6 hours. Gene Symbol Chip ID Fold Change Genbank UniGene Description CCL20 205476_at 421.7 NM_004591 Hs.75498 chemokine (C-C motif) ligand 20 CCL4 204103_at 351.6 NM_002984 Hs.75703 chemokine (C-C motif) ligand 4 BCL2A1 205681_at 228.6 NM_004049 Hs.227817 BCL2-related protein A1 ISG20 204698_at 202.1 NM_002201 Hs.459265 interferon stimulated exonuclease gene 20kDa CCL8 214038_at 139.1 AI984980 Hs.271387 chemokine (C-C motif) ligand 8 CXCL1 204470_at 115.1 NM_001511 Hs.789 chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) IL7R 205798_at 111.6 NM_002185 Hs.362807 interleukin 7 receptor ; interleukin 7 receptor BCL3 204908_s_at 104.2 NM_005178 Hs.31210 B-cell CLL/lymphoma 3 GBP1 202269_x_at 99.99 BC002666 Hs.62661 guanylate binding protein 1, interferon-inducible, 67kDa SLAMF7 219159_s_at 98.3 NM_021181 Hs.517265 SLAM family member 7 CXCL10 204533_at 93.05 NM_001565 Hs.413924 chemokine (C-X-C motif) ligand 10 IL8 211506_s_at 91.31 AF043337 Hs.624 interleukin 8 IL8 202859_x_at 85.26 NM_000584 Hs.624 interleukin 8 THBS1 201110_s_at 84.5 NM_003246 Hs.164226 thrombospondin 1 EBI3 219424_at 82.87 NM_005755 Hs.501452 Epstein-Barr virus induced gene 3 SAT 213988_s_at 76.47 BE971383 Hs.28491 spermidine/spermine N1-acetyltransferase USP18 219211_at 76.22 NM_017414 Hs.38260 ubiquitin specific peptidase 18 LAMP3 205569_at 75.89 NM_014398 Hs.518448 lysosomal-associated membrane protein 3 CCL2 216598_s_at 71.33 S69738 Hs.303649 chemokine -

Downloaded from Consensuspathdb Collection We Defined High-Confidence MTHFD2-Interacting Pro- (
Koufaris and Nilsson Cancer & Metabolism (2018) 6:12 https://doi.org/10.1186/s40170-018-0185-4 RESEARCH Open Access Protein interaction and functional data indicate MTHFD2 involvement in RNA processing and translation Costas Koufaris1,2,3 and Roland Nilsson1,2,3* Abstract Background: The folate-coupled metabolic enzyme MTHFD2 is overexpressed in many tumor types and required for cancer cell proliferation, and is therefore of interest as a potential cancer therapeutic target. However, recent evidence suggests that MTHFD2 has a non-enzymatic function which may underlie the dependence of cancer cells on this protein. Understanding this non-enzymatic function is important for optimal targeting of MTHFD2 in cancer. Methods: To identify potential non-enzymatic functions of MTHFD2, we defined its interacting proteins using co-immunoprecipitation and mass spectrometry and integrated this information with large-scale co-expression analysis, protein dynamics, and gene expression response to MTHFD2 knockdown. Results: We found that MTHFD2 physically interacts with a set of nuclear proteins involved in RNA metabolism and translation, including components of the small ribosomal subunit and multiple members of the RNA-processing hnRNP family. Interacting proteins were also in general co-expressed with MTHFD2 in experiments that stimulate or repress proliferation, suggesting a close functional relationship. Also, unlike other folate one-carbon enzymes, the MTHFD2 protein has a short half-life and responds rapidly to serum. Finally, shRNA against MTHFD2 depletes several of its interactors and yields an overall transcriptional response similar to targeted inhibition of certain ribosomal subunits. Conclusions: Taken together, our findings suggest a novel function of MTHFD2 in RNA metabolism and translation. -

Mitochondrial Methylenetetrahydrofolate
Published OnlineFirst June 22, 2015; DOI: 10.1158/1541-7786.MCR-15-0117 Perspective Molecular Cancer Research Mitochondrial Methylenetetrahydrofolate Dehydrogenase (MTHFD2) Overexpression Is Associated with Tumor Cell Proliferation and Is a Novel Target for Drug Development Philip M. Tedeschi1, Alexei Vazquez2, John E. Kerrigan3, and Joseph R. Bertino4 Abstract Rapidly proliferating tumors attempt to meet the demands for sensitive to treatment with MTX. A key enzyme upregulated in nucleotide biosynthesis by upregulating folate pathways that rapidly proliferating tumors but not in normal adult cells is the provide the building blocks for pyrimidine and purine biosyn- mitochondrial enzyme methylenetetrahydrofolate dehydroge- thesis. In particular, the key role of mitochondrial folate enzymes nase (MTHFD2). This perspective outlines the rationale for spe- in providing formate for de novo purine synthesis and for provid- cific targeting of MTHFD2 and compares known and generated ing the one-carbon moiety for thymidylate synthesis has been crystal structures of MTHFD2 and closely related enzymes as a recognized in recent studies. We have shown a significant corre- molecular basis for developing therapeutic agents against lation between the upregulation of the mitochondrial folate MTHFD2. Importantly, the development of selective inhibitors enzymes, high proliferation rates, and sensitivity to the folate of mitochondrial methylenetetrahydrofolate dehydrogenase is antagonist methotrexate (MTX). Burkitt lymphoma and diffuse expected to have substantial activity, and this perspective supports large-cell lymphoma tumor specimens have the highest levels of the investigation and development of MTHFD2 inhibitors for mitochondrial folate enzyme expression and are known to be anticancer therapy. Mol Cancer Res; 13(10); 1361–6. Ó2015 AACR. Introduction this activity in embryos was shown by knocking out this gene (nmdmc) in mice. -

A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression
Supplementary Materials A Meta-Analysis of the Effects of High-LET Ionizing Radiations in Human Gene Expression Table S1. Statistically significant DEGs (Adj. p-value < 0.01) derived from meta-analysis for samples irradiated with high doses of HZE particles, collected 6-24 h post-IR not common with any other meta- analysis group. This meta-analysis group consists of 3 DEG lists obtained from DGEA, using a total of 11 control and 11 irradiated samples [Data Series: E-MTAB-5761 and E-MTAB-5754]. Ensembl ID Gene Symbol Gene Description Up-Regulated Genes ↑ (2425) ENSG00000000938 FGR FGR proto-oncogene, Src family tyrosine kinase ENSG00000001036 FUCA2 alpha-L-fucosidase 2 ENSG00000001084 GCLC glutamate-cysteine ligase catalytic subunit ENSG00000001631 KRIT1 KRIT1 ankyrin repeat containing ENSG00000002079 MYH16 myosin heavy chain 16 pseudogene ENSG00000002587 HS3ST1 heparan sulfate-glucosamine 3-sulfotransferase 1 ENSG00000003056 M6PR mannose-6-phosphate receptor, cation dependent ENSG00000004059 ARF5 ADP ribosylation factor 5 ENSG00000004777 ARHGAP33 Rho GTPase activating protein 33 ENSG00000004799 PDK4 pyruvate dehydrogenase kinase 4 ENSG00000004848 ARX aristaless related homeobox ENSG00000005022 SLC25A5 solute carrier family 25 member 5 ENSG00000005108 THSD7A thrombospondin type 1 domain containing 7A ENSG00000005194 CIAPIN1 cytokine induced apoptosis inhibitor 1 ENSG00000005381 MPO myeloperoxidase ENSG00000005486 RHBDD2 rhomboid domain containing 2 ENSG00000005884 ITGA3 integrin subunit alpha 3 ENSG00000006016 CRLF1 cytokine receptor like -

Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-Trnas
Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Minton, Denise R. et al. "Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs." Molecular Cell 69 (2018): 610-621 © 2018 The Author(s) As Published 10.1016/j.molcel.2018.01.024 Publisher Elsevier BV Version Author's final manuscript Citable link https://hdl.handle.net/1721.1/124709 Terms of Use Creative Commons Attribution-NonCommercial-NoDerivs License Detailed Terms http://creativecommons.org/licenses/by-nc-nd/4.0/ HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Mol Cell Manuscript Author . Author manuscript; Manuscript Author available in PMC 2019 February 15. Published in final edited form as: Mol Cell. 2018 February 15; 69(4): 610–621.e5. doi:10.1016/j.molcel.2018.01.024. Serine Catabolism by SHMT2 is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl tRNAs Denise R. Minton1,5, Minwoo Nam1,5, Daniel J. McLaughlin1,5, Jong Shin1, Erol C. Bayraktar2, Samantha W. Alvarez1, Vladislav O. Sviderskiy1, Thales Papagiannakopoulos1, David M. Sabatini3, Kıvanç Birsoy2, and Richard Possemato1,4 1Department of Pathology, New York University School of Medicine, New York, New York, 10016 USA. Laura & Isaac Perlmutter Cancer Center, NYU School of Medicine, New York, NY 10016, USA 2Laboratory of Metabolic Regulation and Genetics, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA 3Whitehead Institute for Biomedical Research, Nine Cambridge Center, Cambridge, Massachusetts 02142, USA; Howard Hughes Medical Institute and Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA; The David H. -
![Viewed in [13])](https://docslib.b-cdn.net/cover/3109/viewed-in-13-5053109.webp)
Viewed in [13])
Pelleri et al. BMC Medical Genomics (2014) 7:63 DOI 10.1186/s12920-014-0063-z RESEARCH ARTICLE Open Access Integrated differential transcriptome maps of Acute Megakaryoblastic Leukemia (AMKL) in children with or without Down Syndrome (DS) Maria Chiara Pelleri1, Allison Piovesan1, Maria Caracausi1, Anna Concetta Berardi2, Lorenza Vitale1* and Pierluigi Strippoli1,3 Abstract Background: The incidence of Acute Megakaryoblastic Leukemia (AMKL) is 500-fold higher in children with Down Syndrome (DS) compared with non-DS children, but the relevance of trisomy 21 as a specific background of AMKL in DS is still an open issue. Several Authors have determined gene expression profiles by microarray analysis in DS and/or non-DS AMKL. Due to the rarity of AMKL, these studies were typically limited to a small group of samples. Methods: We generated integrated quantitative transcriptome maps by systematic meta-analysis from any available gene expression profile dataset related to AMKL in pediatric age. This task has been accomplished using a tool recently described by us for the generation and the analysis of quantitative transcriptome maps, TRAM (Transcriptome Mapper), which allows effective integration of data obtained from different experimenters, experimental platforms and data sources. This allowed us to explore gene expression changes involved in transition from normal megakaryocytes (MK, n=19) to DS (n=43) or non-DS (n=45) AMKL blasts, including the analysis of Transient Myeloproliferative Disorder (TMD, n=20), a pre-leukemia condition. Results: We propose a biological model of the transcriptome depicting progressive changes from MK to TMD and then to DS AMKL. The data indicate the repression of genes involved in MK differentiation, in particular the cluster on chromosome 4 including PF4 (platelet factor 4) and PPBP (pro-platelet basic protein); the gene for the mitogen-activated protein kinase MAP3K10 and the thrombopoietin receptor gene MPL. -

Structural and Functional Studies of Proteins of Medical Relevance
! "# $ #%## & ' ( ) * ( + !% , %- % . ( ( / % . ( % (/ %0 / / ( % &01 ( / / 2 3 %. (/ %- -/ &01 %. &01 / &01 ( %. / (/ (/ &01 ( / --%- --- &01 / &01 %4 !&01 / / / 5% &01 ( / -6%0 / / %- / &01 ( 6/ &01 % &01" %&01" ( 5/ % 6- &01"( / %0 &01" % 0 %0 ( / %1/( / ( %- 6--/ (78 9,"( (/ %0 ( %0 / %0/ / (/ % !"!" # "# $ :;; %%; < = : ::: >#!$$ -2?8$? 88?8#?8" -2?8$? 88?8#?$? ( #!? STRUCTURAL AND FUNCTIONAL STUDIES OF PROTEINS OF MEDICAL RELEVANCE Robert Gustafsson Structural and functional studies of proteins of medical relevance Protein-ligand complexes in cancer and novel structural folds in bacteria Robert Gustafsson ©Robert Gustafsson, Stockholm University 2018 ISBN print 978-91-7797-097-2 ISBN PDF 978-91-7797-098-9 Cover picture by Robert Gustafsson and Elisabeth Westergren Printed in Sweden by Universitetsservice US-AB, Stockholm 2018 Distributor: Department of Biochemistry and Biophysics, Stockholm University To my family, and Elisabeth, with all my love List of Publications The following publications and -

Downloaded from and Summarized As
bioRxiv preprint doi: https://doi.org/10.1101/524561; this version posted January 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Transcriptomic correlates of electrophysiological and morphological diversity 2 within and across neuron types 3 Short title: Transcriptomic correlates of neuronal diversity 4 Claire Bomkamp*1,2, Shreejoy J. Tripathy*1,3,5, Carolina Bengtsson Gonzales4, Jens Hjerling-Leffler4, 5 Ann Marie Craig1,2, Paul Pavlidis1,3 6 *These authors contributed equally 7 1 Department of Psychiatry, University of British Columbia, Vancouver BC, Canada 8 2Djavad Mowafaghian Centre for Brain Health, University of British Columbia, Vancouver BC, Canada 9 3Michael Smith Laboratories, University of British Columbia, Vancouver BC, Canada 10 4Laboratory of Molecular Neurobiology, Department of Medical Biochemistry and Biophysics, 11 Karolinska Institutet, SE-17177 Stockholm, Sweden 12 5Present address: Krembil Centre for Neuroinformatics, Centre for Addiction and Mental Health and 13 Department of Psychiatry, University of Toronto, Toronto ON, Canada 14 Abstract 15 In order to further our understanding of how gene expression contributes to key functional properties of 16 neurons, we combined publicly accessible gene expression, electrophysiology, and morphology 17 measurements to identify cross-cell type correlations between these data modalities. Building on our 18 previous work using a similar approach, we distinguished between correlations which were “class- 1 bioRxiv preprint doi: https://doi.org/10.1101/524561; this version posted January 18, 2019.