Unusual Accelerated Rate of Deletions and Insertions in Toxin Genes in The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BIOLOGICAL SURVEY of KANGAROO ISLAND SOUTH AUSTRALIA in NOVEMBER 1989 and 1990
A BIOLOGICAL SURVEY OF KANGAROO ISLAND SOUTH AUSTRALIA IN NOVEMBER 1989 and 1990 Editors A. C. Robinson D. M. Armstrong Biological Survey and Research Section Heritage and Biodiversity Division Department for Environment, Heritage and Aboriginal Affairs, South Australia 1999 i Kangaroo Island Biological Survey The Biological Survey of Kangaroo Island, South Australia was carried out with the assistance of funds made available by, the Commonwealth of Australia under the 1989-90 National Estate Grants Programs and the State Government of South Australia. The views and opinions expressed in this report are those of the authors and do not necessarily represent the views or policies of the Australian Heritage Commission or the State Government of South Australia. The report may be cited as: Robinson, A. C. & Armstrong, D. M. (eds) (1999) A Biological Survey of Kangaroo Island, South Australia, 1989 & 1990. (Heritage and Biodiversity Section, Department for Environment, Heritage and Aboriginal Affairs, South Australia). Copies of the report may be accessed in the library: Environment Australia Department for Environment, Heritage and Aboriginal Affairs GPO Box 636 or 1st Floor, Roma Mitchell House CANBERRA ACT 2601 136 North Terrace, ADELAIDE SA 5000 EDITORS A.C. Robinson, D.M. Armstrong, Biological Survey and Research, Heritage &Biodiversity Section, Department for Environment Heritage and Aboriginal Affairs PO Box 1047 ADELAIDE 5001 AUTHORS D M Armstrong, P.J.Lang, A C Robinson, Biological Survey and Research, Heritage &Biodiversity Section, Department for Environment, Heritage and Aboriginal Affairs PO Box 1047 ADELAIDE 5001 N Draper, Australian Cultural Heritage Management Pty Ltd, 53 Hackney Rd. HACKNEY, SA 5069 G Carpenter, Biodiversity Monitoring and Evaluation, Heritage &Biodiversity Section, Department for Environment Heritage and Aboriginal Affairs. -

Structure±Function Properties of Venom Components from Australian Elapids
PERGAMON Toxicon 37 (1999) 11±32 Review Structure±function properties of venom components from Australian elapids Bryan Grieg Fry * Peptide Laboratory, Centre for Drug Design and Development, University of Queensland, St. Lucia, Qld, 4072, Australia Received 9 December 1997; accepted 4 March 1998 Abstract A comprehensive review of venom components isolated thus far from Australian elapids. Illustrated is that a tremendous structural homology exists among the components but this homology is not representative of the functional diversity. Further, the review illuminates the overlooked species and areas of research. # 1998 Elsevier Science Ltd. All rights reserved. 1. Introduction Australian elapids are well known to be the most toxic in the world, with all of the top ten and nineteen of the top 25 elapids with known LD50s residing exclusively on this continent (Broad et al., 1979). Thus far, three main types of venom components have been characterised from Australian elapids: prothrombin activating enzymes; lipases with a myriad of potent activities; and powerful peptidic neurotoxins. Many species have the prothrombin activating enzymes in their venoms, the vast majority contain phospholipase A2s and all Australian elapid venoms are suspected to contain peptidic neurotoxins. In addition to the profound neurological eects such as disorientation, ¯accid paralysis and respiratory failure, characteristic of bites by many species of Australian elapids is hemorrhaging and incoagulable blood. As a result, these elapids can be divided into two main classes: species with procoagulant venom (Table 1) and species with non-procoagulant venoms (Table 2) (Tan and * Author to whom correspondence should be addressed. 0041-0101/98/$ - see front matter # 1998 Elsevier Science Ltd. -

Austrelaps Labialis Pygmy Copperhead
REPTILE Austrelaps labialis Pygmy Copperhead AUS SA AMLR Endemism Residency - - V State Resident Photo: © Tony Robinson Conservation Significance Endemic to SA. The AMLR distribution is disjunct, isolated from other extant occurrences within SA. Within the AMLR the species’ relative area of occupancy is classified as ‘Extremely Restricted’.3 Although rated Vulnerable in The Action Plan for Australian Reptiles the Pygmy Copperhead has no threatened status under either State or Federal government legislation (Cogger et al. 1993).1 Further information: Biodiversity Conservation Unit, Adelaide Region Phone: (61 8) 8336 0901 Fax: (61 8) 8336 0999 http://www.environment.sa.gov.au/ Department for Environment and Heritage FIS 90346 May 2008 Prepared as part of the Regional Recovery Plan for Threatened Species and Ecological Communities of Adelaide and the Mount Lofty Ranges, South Australia 2009 - 2014 References Ovoviviparous. Pregnant females can be found from Note: In some cases original reference sources are not mid-spring to late summer (November-March) and included in this list, however they can be obtained from the wild caught snakes have given birth in mid-summer reference from which the information has been sourced (the (February) (Jenner 1995; Read and Bedford 1991; reference cited in superscript). 5 Shine 1987a). Mean litter sizes is 7.4 (Shine 1987a). 1 Armstrong, D. M., Croft, S. N. and Foulkes, J. N. (2003). A Biological Survey of the Southern Mount Lofty Ranges, South Aboriginal Significance Australia, 2000-2001. Department for Environment and Post-1983 records indicate the AMLR distribution Heritage, South Australia. occurs in Ngarrindjeri, Kaurna and Peramangk Nations.3 2 Cogger, H. -

A Clinician's Guide to Australian Venomous Bites and Stings
Incorporating the updated CSL Antivenom Handbook Bites and Stings Australian Venomous Guide to A Clinician’s A Clinician’s Guide to DC-3234 Australian Venomous Bites and Stings Incorporating the updated CSL Antivenom Handbook Associate Professor Julian White Associate Professor Principal author Principal author Principal author Associate Professor Julian White The opinions expressed are not necessarily those of bioCSL Pty Ltd. Before administering or prescribing any prescription product mentioned in this publication please refer to the relevant Product Information. Product Information leafl ets for bioCSL’s antivenoms are available at www.biocsl.com.au/PI. This handbook is not for sale. Date of preparation: February 2013. National Library of Australia Cataloguing-in-Publication entry. Author: White, Julian. Title: A clinician’s guide to Australian venomous bites and stings: incorporating the updated CSL antivenom handbook / Professor Julian White. ISBN: 9780646579986 (pbk.) Subjects: Bites and stings – Australia. Antivenins. Bites and stings – Treatment – Australia. Other Authors/ Contributors: CSL Limited (Australia). Dewey Number: 615.942 Medical writing and project management services for this handbook provided by Dr Nalini Swaminathan, Nalini Ink Pty Ltd; +61 416 560 258; [email protected]. Design by Spaghetti Arts; +61 410 357 140; [email protected]. © Copyright 2013 bioCSL Pty Ltd, ABN 26 160 735 035; 63 Poplar Road, Parkville, Victoria 3052 Australia. bioCSL is a trademark of CSL Limited. bioCSL understands that clinicians may need to reproduce forms and fl owcharts in this handbook for the clinical management of cases of envenoming and permits such reproduction for these purposes. However, except for the purpose of clinical management of cases of envenoming from bites/stings from Australian venomous fauna, no part of this publication may be reproduced by any process in any language without the prior written consent of bioCSL Pty Ltd. -

Dynamic Evolution of Australian Elapid Snake Toxins
Toxins 2013, 5, 2621-2655; doi:10.3390/toxins5122621 OPEN ACCESS toxins ISSN 2072-6651 www.mdpi.com/journal/toxins Article Venom Down Under: Dynamic Evolution of Australian Elapid Snake Toxins Timothy N. W. Jackson 1,2,†, Kartik Sunagar 3,4,†, Eivind A. B. Undheim 1,2, Ivan Koludarov 1, Angelo H. C. Chan 1, Kate Sanders 5, Syed A. Ali 1,2,6, Iwan Hendrikx 1, Nathan Dunstan 7 and Bryan G. Fry 1,2,* 1 Venom Evolution Lab, School of Biological Sciences, The University of Queensland, St. Lucia QLD 4072, Australia 2 Institute for Molecular Bioscience, The University of Queensland, St. Lucia QLD 4072, Australia 3 Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, Rua do Campo Alegre, 4169-007, Porto, Portugal 4 CIIMAR/CIMAR-Interdisciplinary Centre of Marine and Environmental Research, University of Porto, Rua dos Bragas 289, P 4050-123 Porto, Portugal 5 School of Earth and Environmental Sciences, University of Adelaide, SA 5005, Australia 6 HEJ Research Institute of Chemistry, International Centre for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi-75270, Pakistan 7 Venom Supplies Pty Ltd, Stonewell Rd, Tanunda SA 5352, Australia † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +61-400-193-182. Received: 14 September 2013; in revised form: 13 December 2013 / Accepted: 16 December 2013 / Published: 18 December 2013 Abstract: Despite the unparalleled diversity of venomous snakes in Australia, research has concentrated on a handful of medically significant species and even of these very few toxins have been fully sequenced. -

Denisonia Hydrophis Parapistocalamus Toxicocalamus Disteira Kerilia Pelamis Tropidechis Drysdalia Kolpophis Praescutata Vermicella Echiopsis Lapemis
The following is a work in progress and is intended to be a printable quick reference for the venomous snakes of the world. There are a few areas in which common names are needed and various disputes occur due to the nature of such a list, and it will of course be continually changing and updated. And nearly all species have many common names, but tried it simple and hopefully one for each will suffice. I also did not include snakes such as Heterodon ( Hognoses), mostly because I have to draw the line somewhere. Disclaimer: I am not a taxonomist, that being said, I did my best to try and put together an accurate list using every available resource. However, it must be made very clear that a list of this nature will always have disputes within, and THIS particular list is meant to reflect common usage instead of pioneering the field. I put this together at the request of several individuals new to the venomous endeavor, and after seeing some very blatant mislabels in the classifieds…I do hope it will be of some use, it prints out beautifully and I keep my personal copy in a three ring binder for quick access…I honestly thought I knew more than I did…LOL… to my surprise, I learned a lot while compiling this list and I hope you will as well when you use it…I also would like to thank the following people for their suggestions and much needed help: Dr.Wolfgang Wuster , Mark Oshea, and Dr. Brian Greg Fry. -

9 Appendices
Smith Bay Ecological Assessment 9 APPENDICES Appendix 1. Compliance and Legislation summary. Environment Protection and Biodiversity Conservation Act 1999 The Environment Protection and Biodiversity Conservation Act 1999 (EPBC Act) provides a legal framework to protect and manage nationally and internationally important flora, fauna, ecological communities and heritage places – defined in the Act as ‘matters of national environmental significance’. The nine matters of national environmental significance protected under the Act are: World Heritage properties National Heritage places wetlands of international importance (listed under the Ramsar Convention) listed threatened species and ecological communities migratory species protected under international agreements Commonwealth marine areas the Great Barrier Reef Marine Park nuclear actions (including uranium mines) a water resource, in relation to coal seam gas development and large coal mining development. DRAFT Any action that has, will have, or is likely to have a significant impact on matters of national environmental significance requires referral under the EPBC Act. Substantial penalties apply for undertaking an action that has, will have or is likely to have significant impact on a matter of national environmental significance without approval. This report is focused on listed threatened species and ecological communities which are recognised as a matter of national environmental significance. Consequently, any action that is likely to have a significant impact on listed threatened species and ecological communities under the EPBC Act must be referred to the Minister and undergo an environmental assessment and approval process. The EPBC Act Significant Impact Guidelines (Commonwealth of Australia 2013) provide overarching guidance on determining whether an action is likely to have a significant impact on a matter of national environmental significance. -

Snake Venomics and Antivenomics: Proteomic Tools in the Design and Control of Antivenoms for the Treatment of Snakebite Envenoming
JOURNAL OF PROTEOMICS 72 (2009) 165– 182 available at www.sciencedirect.com www.elsevier.com/locate/jprot Review Snake venomics and antivenomics: Proteomic tools in the design and control of antivenoms for the treatment of snakebite envenoming José María Gutiérreza,⁎, Bruno Lomontea, Guillermo Leóna, Alberto Alape-Giróna,b,c, Marietta Flores-Díaza, Libia Sanzd, Yamileth Anguloa, Juan J. Calveted aInstituto Clodomiro Picado, Facultad de Microbiología, Universidad de Costa Rica, San José, Costa Rica bDepartamento de Bioquímica, Escuela de Medicina, Universidad de Costa Rica, San José, Costa Rica cCentro de Investigación en Estructuras Microscópicas, Universidad de Costa Rica, San José, Costa Rica dInstituto de Biomedicina de Valencia, CSIC, Valencia, Spain ARTICLE DATA ABSTRACT Article history: Snakebite envenoming represents a neglected tropical disease that has a heavy public Received 23 December 2008 health impact, particularly in Asia, Africa and Latin America. A global initiative, aimed at Accepted 7 January 2009 increasing antivenom production and accessibility, is being promoted by the World Health Organization and others. This work discusses several aspects of antivenom manufacture Keywords: and control in which the proteomic analysis of snake venoms, for which the term ‘snake Proteomics of snake venoms venomics’ has been coined, might play a relevant supporting role. Snake venomics has Venomics already shown its usefulness for generating knowledge at different levels (ontogenetic, Snake venoms individual, and geographic) on inter- and intraspecies venom variability. This information Antivenoms has applications for the quality control of venom preparations used in antivenom Antivenomics manufacture. Moreover, the design of the best venom mixtures for immunization, aimed Preclinical testing at increasing the effectiveness of antivenoms, may also be guided by venom proteome analysis, including molecular studies of the cross-reactivity of antivenoms and heterologous venoms through a recently developed methodological approach termed ‘antivenomics’. -

HCN Proposed Reptiles Species List
HCN Proposed Reptiles Species List All content in this publication is owned by the Herpetocultural Cooperative of NSW Complied by the HCN Committee on behalf of our members and community. Contributors to this document include representatives from: Australian Herpetological Society, Central Coast Herpetological Society, Hawkesbury Herpetological Society, Illawarra Reptile Society, Macarthur Herpetological Society, North Coast Herpetological Group, Shoalhaven Reptile Club, Turtles R Us, DoLittle Farms, Flora & Fauna Management Services, Wildexpos, and other private reptile keepers. 2 Contents: 1. Nomenclateral changes & species division 4 Turtles 4 Lizards, geckos 4 Lizards, skinks 7 Lizards, dragons 9 Lizards, goannas 11 Snakes, pythons 12 Snakes elapids 12 2. List of Coded Species 13 3. Proposed method for changes to assignment/addition of taxa to Reptile 19 License categories Addition of new species NOT native to NSW 19 Addition of new species that occur in natural populations in NSW 20 Changes in category species 20 4. Current allocation of venomous snakes to Reptile Keeper Categories 21 5. Proposal for additions of several species to current licensing categories 23 Partial list of lizards & snakes for which applications to hold or import may 25 occur Pygopods 25 Skins 25 Dragons 26 Goannas 26 Pythons 27 elapids 27 Draft species Risk Assessment Tools 29 Black spined Nobbi Dragon 29 Slater’s Ring-tailed Dragon 32 Goldfield’s Ring-tailed Dragon 35 White-lipped Two-lined Dragon 38 Arnhem Two-lined Dragon 41 Lally’s Two-lined Dragon 44 Thorny Devil 47 North-west Red-faced Turtle 49 Northern Yellow-faced Turtle 54 Painted Short-necked Turtle 57 3 The HCN notes that the proposed classification of reptiles to Reptile Keeper Classes makes no changes to the existing listings other than to remove 11 species from Class 1 to a new Code- regulated category. -
Table 7: Species Changing IUCN Red List Status (2017-2018)
IUCN Red List version 2018-2: Table 7 Last Updated: 14 November 2018 Table 7: Species changing IUCN Red List Status (2017-2018) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2017 (IUCN Red List version 2017-3) and 2018 (IUCN Red List version 2018-2) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2017) List (2018) change version Category -

A Review and Database of Snake Venom Proteomes
toxins Review A Review and Database of Snake Venom Proteomes Theo Tasoulis and Geoffrey K. Isbister * ID Clinical Toxicology Research Group, University of Newcastle, Newcastle 2298, Australia; [email protected] * Correspondence: [email protected]; Tel.: +612-4921-1211 Academic Editor: Eivind Undheim Received: 1 September 2017; Accepted: 15 September 2017; Published: 18 September 2017 Abstract: Advances in the last decade combining transcriptomics with established proteomics methods have made possible rapid identification and quantification of protein families in snake venoms. Although over 100 studies have been published, the value of this information is increased when it is collated, allowing rapid assimilation and evaluation of evolutionary trends, geographical variation, and possible medical implications. This review brings together all compositional studies of snake venom proteomes published in the last decade. Compositional studies were identified for 132 snake species: 42 from 360 (12%) Elapidae (elapids), 20 from 101 (20%) Viperinae (true vipers), 65 from 239 (27%) Crotalinae (pit vipers), and five species of non-front-fanged snakes. Approximately 90% of their total venom composition consisted of eight protein families for elapids, 11 protein families for viperines and ten protein families for crotalines. There were four dominant protein families: phospholipase A2s (the most common across all front-fanged snakes), metalloproteases, serine proteases and three-finger toxins. There were six secondary protein families: cysteine-rich secretory proteins, L-amino acid oxidases, kunitz peptides, C-type lectins/snaclecs, disintegrins and natriuretic peptides. Elapid venoms contained mostly three-finger toxins and phospholipase A2s and viper venoms metalloproteases, phospholipase A2s and serine proteases. Although 63 protein families were identified, more than half were present in <5% of snake species studied and always in low abundance. -
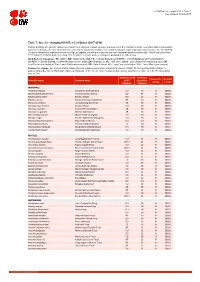
Table 7: Species Changing IUCN Red List Status (2017-2018)
IUCN Red List version 2018-1: Table 7 Last Updated: 05 July 2018 Table 7: Species changing IUCN Red List Status (2017-2018) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2017 (IUCN Red List version 2017-3) and 2018 (IUCN Red List version 2018-1) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2017) List (2018) change version Category