Artocarpus Integer, Moraceae), and Its Wild Relative Bangkong (Artocarpus Integer Var
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LSS Bukit Selambau Dilancar
Headline LSS Bukit Selambau dilancar MediaTitle Sinar Harian Date 01 Apr 2019 Language Malay Circulation 140,000 Readership 420,000 Section Selangor & KL Page No 37 ArticleSize 438 cm² Journalist SITI ZUBAIDAH PR Value RM 22,414 LSS Bukit Selambau dilancar Bawa manfaat bandar Sungai Petani itu. “Kita baru sahaja selesai info kepada menyempurnakan pro- penduduk, jadi gram terbesar dan berpres- Projek LLS di Bukit tij Pameran Aeroangkasa Selambau itu dijangka mercu tanda dan Maritim Antarabangsa mula beroperasi pada Langkawi 2019 (LIMA’19) hujung tahun 2020 yang berjalan lancar, anta- SITI ZUBAIDAH ZAKARAYA ra lain, hasil sokongan dan kerjasama TNB dengan ngan tarikh sasaran opera- SUNGAI PETANI menyiapkan sistem auto- si komersial pada 31 masi sepenuhnya peman- Disember 2020. tauan bekalan elektrik di “TNB yakin ini meru- rojek Solar Berskala Pulau Langkawi. pakan lokasi tepat berikut- P Besar (LSS) Tenaga “Saya difahamkan an Kedah antara negeri Nasional Berhad pemasangan sistem ini yang banyak menerima (TNB) bernilai RM180 juta membolehkan TNB cahaya matahari selepas bakal memanfaatkan pen- memantau dengan lebih Perlis berbanding negeri duduk Bukit Selambau, rapi lagi bekalan elektrik di Mukhriz menurunkan tandatangan sebagai gimik pelancaran Majlis Pecah Tanah LSS di Pekan Bukit lain di semenanjung,” ka- sekaligus membangunkan seluruh Pulau Langkawi Selambau, semalam. tanya. kawasan pinggir bandar dan bertindak pantas Beliau percaya, projek berkenaan. apabila ada gangguan dan bertuah kerana bakal dide- naga suria ini juga amat tama TNB dianugerahkan berkenaan sedikit sebanyak Menteri Besar Kedah, sebagainya, dan tempoh dahkan dengan perkem- sesuai di Kedah, kerana kita LSS oleh Suruhanjaya Te- memberi kesan limpahan Datuk Seri Mukhriz Ma- masa pemulihan dapat bangan teknologi masa antara negeri yang mene- naga adalah melalui proses kepada rakyat menerusi hathir berkata, projek yang dipendekkan. -

LK Audited Per 31 Desember 2016
PT WASKITA KARYA (PERSERO) Tbk PT WASKITA KARYA (PERSERO) Tbk DAN ENTITAS ANAK AND SUBSIDIARIES Daftar Isi Halaman/ Table of Contents Pages Surat Pernyataan Direksi Director’s Statement Letter Laporan Auditor Independen Independent Auditor’s Report Laporan Keuangan Konsolidasian Consolidated Financial Statements Untuk Tahun-tahun yang Berakhir pada For the Years Ended Tanggal 31 Desember 2016 dan 2015 December 31, 2016 and 2015 Laporan Posisi Keuangan Konsolidasian 1 Consolidated Statements of Financial Position Laporan Laba Rugi dan Penghasilan 3 Consolidated Statements of Profit or Loss and Komprehensif Lain Konsolidasian Other Comprehensive Income Laporan Perubahan Ekuitas Konsolidasian 4 Consolidated Statements of Changes in Equity Laporan Arus Kas Konsolidasian 5 Consolidated Statements of Cash Flows Catatan Atas Laporan Keuangan 6 Notes to the Consolidated the Financial Konsolidasian Statements PT WASKITA KARYA (PERSERO) Tbk PT WASKITA KARYA (PERSERO) Tbk DAN ENTITAS ANAK AND SUBSIDIARIES LAPORAN POSISI KEUANGAN CONSOLIDATED STATEMENTS OF KONSOLIDASIAN FINANCIAL POSITION 31 Desember 2016 dan 2015 December 31, 2016 and 2015 (Dalam Rupiah Penuh) (In Full of Rupiah) Catatan/ 2016 2015 *) Notes Rp Rp ASET ASSETS ASET LANCAR CURRENT ASSETS Kas dan Setara Kas 4, 49, 51, 54 10,653,780,768,186 5,511,188,078,778 Cash and Cash Equivalents Investasi Jangka Pendek 5 10,663,933,745 10,663,933,745 Short-Term Investments Piutang Usaha Accounts Receivable Pihak Berelasi 6, 51, 54 1,356,258,910,298 1,174,449,804,260 Related Parties Pihak Ketiga 6 -

Direktori Konstruksi
http://www.bps.go.id http://www.bps.go.id http://www.bps.go.id DIREKTORI PERUSAHAAN KONSTRUKSI 2012 Directory of Construction Establishment 2012 Buku IV (Pulau Kalimantan) Book IV (Pulau Kalimantan) ISBN. 978-979-064-175-7 No. Publikasi / Publication Number : 05340.1005 Katalog BPS / BPS Catalogue : 1305055 Ukuran Buku / Book Size : 21 cm X 29 cm Jumlah Halaman / TotalPage : (xv + 366) halaman / pages Naskah / Manuscript : Subdirektorat Statistik Konstruksi Subdirectorate of Construction Statistics Gambar Kulit / Cover Design : Subdirektorat Statistik Konstruksi Subdirectorate of Construction Statistics Diterbitkan oleh / Published by : http://www.bps.go.id Badan Pusat Statistik, Jakarta, Indonesia BPS-Statistics Indonesia, Jakarta, Indonesia Boleh dikutip dengan menyebut sumbernya May be cited with reference to the sources KATA PENGANTAR Direktori Perusahaan Konstruksi 2012 ini merupakan perbaikan dari Direktori Perusahaan Konstruksi 2011 berdasarkan hasil Survei Updating Direktori Perusahaan Konstruksi Tahun 2012 dan Survei Perusahaan Konstruksi Tahun 2012. Direktori ini merupakan identifikasi perusahaan yang meliputi: KIP, Nama, Alamat, Nomor Telepon, Nomor Faximile, dan Alamat Email Perusahaan. Diharapkan Publikasi ini bermanfaat baik oleh perusahaan bersangkutan maupun konsumen data yang memerlukan untuk kegiatan sehari-harinya. Disamping itu direktori ini diharapkan dapat digunakan juga sebagai kerangka sampel bagi penelitian atau studi-studi khusus selanjutnya. Akhirnya pada kesempatan ini kami mengucapkan terima kasih dan penghargaan kepada semua pihak terutama kepada para Pengusaha dan Pimpinan Perusahaan Jasa Konstruksi yang telah membantu kelancaran pelaksanaan survei tersebut, dan menghimbau di masa mendatang agar dapat memberikan data yang akurat, lengkap dan reliable serta dapat memberikan masukan untuk perbaikan publikasi ini. http://www.bps.go.id Jakarta, September 2012 Kepala Badan Pusat Statistik Republik Indonesia DR. -

Pharmaceutical Importance of Artocarpus Altilis
Human Journals Review Article April 2015 Vol.:3, Issue:1 © All rights are reserved by Chinmay Pradhan et al. A Review on Phytochemistry, Bio-Efficacy, Medicinal and Ethno- Pharmaceutical Importance of Artocarpus altilis Keywords: Artocarpus, Antimicrobial activity, Ethno- pharmacognosy, Phytoconstituent, Therapeutic property ABSTRACT Monalisa Mohanty1 and Chinmay Pradhan2* In recent years, herbal medicine is being the sources of many imperative drugs of modern world. Use of potent medicinal 1 Dhenkanal Autonomous College, Dhenkanal, Odisha, plants like Breadfruit (Artocarpus altilis) against various ailments to reduce the adverse effects of various orthodox India allopathic medicines and detrimental side effects of 2 Laboratory of Microbial Biotechnology, conventional antibiotics has emerged as an evolved technique in pharmaceutical science. The present review emphasizes the Post Graduate Department of Botany antimicrobial potentiality of various parts of A. altilis along Utkal University, Bhubaneswar, Odisha, India with their known therapeutic properties in context of biologically active compounds (phytoconstituents). Research Submission: 9 April 2015 investigations on ethno-pharmacological study of the plant Accepted: 18 April 2015 parts with their nutritional value, multifarious medicinal uses Published: 25 April 2015 and antibacterial effect are being quoted in the present review. This review will provide detailed information to future researchers on phytoconstituent analysis, bioefficacy assessment and ethno-pharmaceutical importance of A. altilis in the field of medical science. www.ijppr.humanjournals.com www.ijppr.humanjournals.com INTRODUCTION In recent years research on the emergence of multiple drug resistance to various human pathogenic bacteria has gained utmost importance all over the world which necessitated a search for new antimicrobial substances from other sources including plants. -
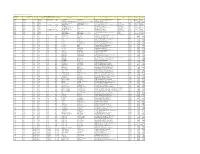
Colgate Palmolive List of Mills As of June 2018 (H1 2018) Direct
Colgate Palmolive List of Mills as of June 2018 (H1 2018) Direct Supplier Second Refiner First Refinery/Aggregator Information Load Port/ Refinery/Aggregator Address Province/ Direct Supplier Supplier Parent Company Refinery/Aggregator Name Mill Company Name Mill Name Country Latitude Longitude Location Location State AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora Agroaceite Extractora Agroaceite Finca Pensilvania Aldea Los Encuentros, Coatepeque Quetzaltenango. Coatepeque Guatemala 14°33'19.1"N 92°00'20.3"W AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora del Atlantico Guatemala Extractora del Atlantico Extractora del Atlantico km276.5, carretera al Atlantico,Aldea Champona, Morales, izabal Izabal Guatemala 15°35'29.70"N 88°32'40.70"O AgroAmerica Agrocaribe Guatemala Agrocaribe S.A Extractora La Francia Guatemala Extractora La Francia Extractora La Francia km. 243, carretera al Atlantico,Aldea Buena Vista, Morales, izabal Izabal Guatemala 15°28'48.42"N 88°48'6.45" O Oleofinos Oleofinos Mexico Pasternak - - ASOCIACION AGROINDUSTRIAL DE PALMICULTORES DE SABA C.V.Asociacion (ASAPALSA) Agroindustrial de Palmicutores de Saba (ASAPALSA) ALDEA DE ORICA, SABA, COLON Colon HONDURAS 15.54505 -86.180154 Oleofinos Oleofinos Mexico Pasternak - - Cooperativa Agroindustrial de Productores de Palma AceiteraCoopeagropal R.L. (Coopeagropal El Robel R.L.) EL ROBLE, LAUREL, CORREDORES, PUNTARENAS, COSTA RICA Puntarenas Costa Rica 8.4358333 -82.94469444 Oleofinos Oleofinos Mexico Pasternak - - CORPORACIÓN -

Kode Dan Data Wilayah Administrasi Pemerintahan Provinsi Kalimantan Barat
KODE DAN DATA WILAYAH ADMINISTRASI PEMERINTAHAN PROVINSI KALIMANTAN BARAT JUMLAH N A M A / J U M L A H LUAS JUMLAH NAMA PROVINSI / K O D E WILAYAH PENDUDUK K E T E R A N G A N (Jiwa) *) KABUPATEN / KOTA KAB KOTA KECAMATAN KELURAHAN D E S A (Km2) 61 KALIMANTAN BARAT 61.01 1 KAB. SAMBAS 19 - 183 6.716,52 562.827 61.01.01 1 Sambas - 18 61.01.01.2001 1 Dalam Kaum 61.01.01.2002 2 Lubuk Dagang 61.01.01.2003 3 Tanjung Bugis 61.01.01.2004 4 Pendawan 61.01.01.2005 5 Pasar Melayu 61.01.01.2006 6 Durian 61.01.01.2007 7 Lorong 61.01.01.2008 8 Jagur 61.01.01.2009 9 Tumuk Manggis 61.01.01.2010 10 Tanjung Mekar 61.01.01.2011 11 Sebayan 61.01.01.2012 12 Kartiasa 61.01.01.2013 13 Saing Rambi 61.01.01.2014 14 Lumbang 61.01.01.2015 15 Sungai Rambah Semanjang Menjadi wil. Kec. Sebawi, Perda No.5/2004. Tebing Batu Menjadi wil. Kec. Sebawi, Perda No.5/2004. Sebawi Menjadi wil. Kec. Sebawi, Perda No.5/2004. Sepuk Tanjung Menjadi wil. Kec. Sebawi, Perda No.5/2004. Sebangun Menjadi wil. Kec. Sebawi, Perda No.5/2004. Sempalai Sebedang Menjadi wil. Kec. Sebawi, Perda No.5/2004. Tempatan Menjadi wil. Kec. Sebawi, Perda No.5/2004. 61.01.01.2023 16 Gapura 61.01.01.2024 17 Sumber Harapan Jirak Menjadi wil. Kec. Sajad , Perda No.5/2004. Tengguli Menjadi wil. -

Consultant's Report of Biodiversity Specialist for Usaid Pid Mission Natural Resources Management in Kalimantan
CONSULTANT'S REPORT OF BIODIVERSITY SPECIALIST FOR USAID PID MISSION NATURAL_ RESOURCES MANAGEMENT IN KALIMANTAN Kathy MacKinnon Ecology Advisor EMDI D~cember 1988 The Biological Importance of Kalimantan Kalimantan, as part of the great·island of·Borneo,theworld's~· third-largest island, is biologically. one of· themost.important-· , ..... -. - --r- areas of Indonesia. Borneo lies within the everwet tropiCS and supports the largest expanse of tropical.~rainforest -in ~the - ~ Indomalayan Realm. It is a main centre of distribution for many genera of the Malesian flora and the Indomalayan fauna. Forest types include mangrove forests, large areas of peat swamp and freshwater non-peaty swamp, the most extensive heath or kerangasforests in the realm, lowland dipterocarp forest, forests on limestone, and variolls montane formations. Geologically and climatically, Borneo has remained stable for the last few thousand years and evolved high species diversity. Table 1 compares species richness and endemism for plants, mammals, birds and reptiles on the Indonesian islands. Borneo and Irian Jaya (New Guinea) score most highly for spedes richness and are the obvious fir.st can:iidates for conservation efforts to protect biological divE:rsi ty in Indonesia. Deforestation and development are proceeding more rapidly in Kalimantan. Kalimantan's need for immediate action makes it the best focus for the USAID natural resources managemElDt project to maintain biological d.iversity. Borneo is very rich in both flora and fauna. Tne illlanJ is the richest unit of the Sundaic subn~gion ",'itn :;mall plot t::-ee diversity as high as fot.::.:lj anywhnre in ~ew (,u1nea or S:ilth America. Borneo, j,'"ith 262 SpeciE!3 of riipteroca=;:s, 1:; i:he centre of distribution :o!' the f,:,.rcily D:!pteroce::-paceae. -

SPAFA Journal
This Page Is Intentionally Left Blank sPA PA Ili~F.ST Vo l. VI No. 1, 1985 ISSN 0125 - 7099 Contents Ancient Beads from Philippine Archaeological Sites by Robert B. Fox and Rey Santiago 4 Theatre and Visual Arts: Some SPAFA objectives Parallelism by Jukka Miettinen 14 The objectives of SPAFA are: The Role of Conservation in Display by Colin Pearson 17 - To promote awareness and appre ciation of the cultural heritage of the The Mor Yao Ceremony to Southeast Asian countries t hrough the Honor the Benevolent Spirits preservation of archaeological and histo by Orawan Banchangsilpa 24 rical artifacts as well as the t raditional arts; West Java's Increasing Involvement in Overseas Treade in the 13th - To help enrich cultural activit ies in and 14th Centuries by E. Edwards the region; McKinnon 28 - To strengthen professio nal compe Preliminary Excavation at Fort tence in the fi elds of archaeology and Canning, Singapore by John Miksic 34 fine arts through sharing of resources and experiences on a regional basis; The Maritime Network in the Indonesian Archipelago in the - To promote better understanding Fourteenth Century by A.B. Lapian 40 among the countries of Southeast Asia through joint programmes in archaeology SPAFA Affairs 46 and fine arts. The Cover The Indonesian wayang (shadow theatre) makes use of flat S/lA/JA buffalo parchment puppets which are played against an upright 171~F.ST screen illuminated from behind. The puppets are lacy silhouettes in profile and are manipulated through three long handles, one for the body and one for each arm. -

Richard B. Primack
Richard B. Primack Department of Biology Boston University Boston, MA 02215 Contact information: Phone: 617-353-2454 Fax: 617-353-6340 E-mail: [email protected] Website: http://www.bu.edu/biology/people/faculty/primack/ Lab Blog: http://primacklab.blogspot.com/ EDUCATION Ph.D. 1976, Duke University, Durham, NC Botany; Advisor: Prof. Janis Antonovics B.A. 1972, Harvard University, Cambridge, MA Biology; magna cum laude; Advisor: Prof. Carroll Wood EMPLOYMENT Boston University Professor (1991–present) Associate Professor (1985–1991) Assistant Professor (1978–1985) Associate Director of Environmental Studies (1996–1998) Faculty Associate: Pardee Center for the Study of the Longer Range Future Biological Conservation, an international journal. Editor (2004–present); Editor-in-Chief (2008-2016). POST-DOCTORAL, RESEARCH, AND SABBATICAL APPOINTMENTS Distinguished Overseas Professor of International Excellence, Northwest Forestry University, Harbin, China. (2014-2017). Visiting Scholar, Concord Museum, Concord, MA (2013) Visiting Professor, Tokyo University, Tokyo, Japan (2006–2007) Visiting Professor for short course, Charles University, Prague (2007) Putnam Fellow, Harvard University, Cambridge, MA (2006–2007) Bullard Fellow, Harvard University, Cambridge, MA (1999–2000) Visiting Researcher, Sarawak Forest Department, Sarawak, Malaysia (1980–1981, 1985– 1990, 1999-2000) Visiting Professor, University of Hong Kong, Hong Kong (1999) Post-doctoral fellow, Harvard University, Cambridge, MA (1980–1981). Advisor: Prof. Peter S. Ashton Post-doctoral fellow, University of Canterbury, Christchurch, New Zealand (1976–1978). Advisor: Prof. David Lloyd 1 HONORS AND SERVICE Environmentalist of the Year Award. Newton Conservators for efforts to protect the Webster Woods. Newton, MA. (2020). George Mercer Award. Awarded by the Ecological Society of America for excellence in a recent research paper lead by a young scientist. -

023 SK Penetapan Status Akreditasi Program Dan Satuan PAUD Dan
FR-AK-11 KEPUTUSAN BADAN AKREDITASI NASIONAL PENDIDIKAN ANAK USIA DINI DAN PENDIDIKAN NONFORMAL NOMOR 023/BAN PAUD PNF/AKR/2017 TENTANG PENETAPAN STATUS AKREDITASI PROGRAM DAN SATUAN PENDIDIKAN ANAK USIA DINI DAN PENDIDIKAN NONFORMAL TAHAP IV TAHUN 2017 BADAN AKREDITASI NASIONAL PENDIDIKAN ANAK USIA DINI DAN PENDIDIKAN NONFORMAL, Menimbang : a. bahwa dalam rangka pelaksanaan Pasal 60 Undang-Undang Nomor 20 Tahun 2003 tentang Sistem Pendidikan Nasional dan Pasal 1 Angka 32 Peraturan Pemerintah Nomor 13 Tahun 2015 tentang Perubahan Kedua Atas Peraturan Pemerintah Republik Indonesia Nomor 19 Tahun 2005 tentang Standar Nasional Pendidikan perlu dilakukan akreditasi terhadap Program dan Satuan Pendidikan Anak Usia Dini dan Pendidikan Nonformal oleh Badan Akreditasi Nasional Pendidikan Anak Usia Dini dan Pendidikan Nonformal; b. bahwa berdasarkan pertimbangan sebagaimana dimaksud dalam huruf a, perlu menetapkan Keputusan Badan Akreditasi Nasional Pendidikan Anak Usia Dini dan Pendidikan Nonformal tentang Penetapan Status Akreditasi Program dan Satuan Pendidikan Anak Usia Dini dan Pendidikan Nonformal Tahap IV Tahun 2017; FR-AK-11 Mengingat : 1. Undang-Undang Nomor 20 Tahun 2003 tentang Sistem Pendidikan Nasional (Lembaran Negara Republik Indonesia Tahun 2003 Nomor 78, Tambahan Lembaran Negara Republik Indonesia Nomor 4301); 2. Peraturan Pemerintah Nomor 19 tahun 2005 tentang Standar Nasional Pendidikan (Lembaran Negara Republik Indonesia Tahun 2015 Nomor 45, Tambahan Lembaran Negara Republik Indonesia Nomor 5670) sebagaimana telah diubah terakhir dengan Peraturan Pemerintah Nomor 13 tahun 2015 (Lembaran Negara Republik Indonesia Tahun 2015 Nomor 45, Tambahan Lembaran Negara Republik Indonesia Nomor 5670); 3. Peraturan Menteri Pendidikan dan Kebudayaan Nomor 52 Tahun 2015 tentang Badan Akreditasi Nasional Pendidikan Anak Usia Dini dan Pendidikan Non Formal (Berita Negara Republik Indonesia Tahun 2015 Nomor 1856); 4. -

Wood for the Trees: a Review of the Agarwood (Gaharu) Trade in Malaysia
WOOD FOR THE TREES : A REVIEW OF THE AGARWOOD (GAHARU) TRADE IN MALAYSIA LIM TECK WYN NOORAINIE AWANG ANAK A REPORT COMMISSIONED BY THE CITES SECRETARIAT Published by TRAFFIC Southeast Asia, Petaling Jaya, Selangor, Malaysia © 2010 The CITES Secretariat. All rights reserved. All material appearing in this publication is copyrighted and may be reproduced with permission. Any reproduction in full or in part of this publication must credit the CITES Secretariat as the copyright owner. This report was commissioned by the CITES Secretariat. The views of the authors expressed in this publication do not however necessarily reflect those of the CITES Secretariat. The geographical designations employed in this publication, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat concerning the legal status of any country, territory, or area, or its authorities, or concerning the definition of its frontiers or boundaries. ~~~~~~~~~~~~~~~~~~ The TRAFFIC symbol copyright and Registered Trademark ownership is held by WWF. TRAFFIC is a joint programme of WWF and IUCN. Suggested citation: Lim Teck Wyn and Noorainie Awang Anak (2010). Wood for trees: A review of the agarwood (gaharu) trade in Malaysia TRAFFIC Southeast Asia, Petaling Jaya, Selangor, Malaysia ISBN 9789833393268 Cover: Specialised agarwood retail shops have proliferated in downtown Kuala Lumpur for the Middle East tourist market Photograph credit: James Compton/TRAFFIC Wood for the trees :A review of the agarwood (gaharu) -

An Audit of Diabetes Control and Management (DRM-ADCM) January – December 2009
Diabetes Registry Malaysia (DRM) Report of An Audit of Diabetes Control and Management (DRM-ADCM) January – December 2009 Editors: Dr Mastura Ismail, Dr Jamaiyah Haniff, Prof Dato’ Wan Mohamed Wan Bebakar With contributions from: Dr Lee Ping Yein, Dr Syed Alwi Syed Abd Rahman, Dr Cheong Ai Theng, Dr Chew Boon How, Dr Sazlina Shariff Ghazali, Dr Zaiton Ahmad, Dr Nafiza Mat Nasir, Dr Sri Wahyu Taher, Dr Mohd Fozi Kamaruddin, Dr Mastura Ismail, Dr Zanariah Hussein, Dr GR Letchuman Ramanathan Diabetes Registry Malaysia (DRM-ADCM) is funded by a grant from the Ministry of Health, Malaysia 1 Published by, Diabetes Registry Malaysia (DRM-ADCM) 1st Floor, MMA House 124 Jalan Pahang 53000 Kuala Lumpur Malaysia Telephone : (603) 40443060/ 3070 Direct Fax : (603) 40443080 Email : [email protected] Website : www.acrm.org.my/adcm This report is copyrighted. Reproduction and dissemination of this report in part or in whole for research, educational or other non-commercial purposes are authorised without any prior written permission from the copyright holders provided the source is fully acknowledged. Important information: The information in this report only represents selected Ministry of Health clinics and therefore is not representative of national statistics. In case of doubts, readers are advised to seek clarification from the editors of this report. Suggested citation: Mastura I, Jamaiyah H, Wan Mohamad WB (Eds). Diabetes Registry Malaysia: Report of An Audit Of Diabetes Control and Management (January- December 2009), Kuala Lumpur 2010 This report