Ideal Chain Conformations and Statistics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

POLYMER STRUCTURE and CHARACTERIZATION Professor
POLYMER STRUCTURE AND CHARACTERIZATION Professor John A. Nairn Fall 2007 TABLE OF CONTENTS 1 INTRODUCTION 1 1.1 Definitions of Terms . 2 1.2 Course Goals . 5 2 POLYMER MOLECULAR WEIGHT 7 2.1 Introduction . 7 2.2 Number Average Molecular Weight . 9 2.3 Weight Average Molecular Weight . 10 2.4 Other Average Molecular Weights . 10 2.5 A Distribution of Molecular Weights . 11 2.6 Most Probable Molecular Weight Distribution . 12 3 MOLECULAR CONFORMATIONS 21 3.1 Introduction . 21 3.2 Nomenclature . 23 3.3 Property Calculation . 25 3.4 Freely-Jointed Chain . 27 3.4.1 Freely-Jointed Chain Analysis . 28 3.4.2 Comment on Freely-Jointed Chain . 34 3.5 Equivalent Freely Jointed Chain . 37 3.6 Vector Analysis of Polymer Conformations . 38 3.7 Freely-Rotating Chain . 41 3.8 Hindered Rotating Chain . 43 3.9 More Realistic Analysis . 45 3.10 Theta (Θ) Temperature . 47 3.11 Rotational Isomeric State Model . 48 4 RUBBER ELASTICITY 57 4.1 Introduction . 57 4.2 Historical Observations . 57 4.3 Thermodynamics . 60 4.4 Mechanical Properties . 62 4.5 Making Elastomers . 68 4.5.1 Diene Elastomers . 68 0 4.5.2 Nondiene Elastomers . 69 4.5.3 Thermoplastic Elastomers . 70 5 AMORPHOUS POLYMERS 73 5.1 Introduction . 73 5.2 The Glass Transition . 73 5.3 Free Volume Theory . 73 5.4 Physical Aging . 73 6 SEMICRYSTALLINE POLYMERS 75 6.1 Introduction . 75 6.2 Degree of Crystallization . 75 6.3 Structures . 75 Chapter 1 INTRODUCTION The topic of polymer structure and characterization covers molecular structure of polymer molecules, the arrangement of polymer molecules within a bulk polymer material, and techniques used to give information about structure or properties of polymers. -

Properties of Two-Dimensional Polymers
Macromolecules 1982,15, 549-553 549 The squared frequencies uA2are obtained from the ei- (4) Flory, P. J. "Statistical Mechanics of Chain Molecules"; In- genvalues Of this matrix described in the text* terscience: New York, 1969; p 315. (5) Volkenstein,M. V. &Configurational Statistics of polymeric for N = 3 and 4 will be found in ref 7. Chains" (translated from the Russian edition by S. N. Ti- masheff and M. J. Timasheff);Interscience: New York, 1963; References and Notes p 450. (1) Weiner, J. H.; Perchak, D. Macromolecules 1981, 14, 1590. (6) Frenkel, J. "Kinetic Theory of Liquids"; Oxford University (2) Weiner, J. H., Macromolecules, preceding paper in this issue. Press: London, 1946, Dover reprint, 1955; pp 474-476. (3) G6, N.; Scheraga, H. A. Macromolecules 1976, 9, 535. (7) Perchak, D. Doctoral Dissertation, Brown University, 1981. Properties of Two-Dimensional Polymers Jan Tobochnik and Itzhak Webman* Department of Mathematics, Rutgers, The State University, New Brunswick, New Jersey 08903 Joel L. Lebowitzt Institute for Advanced Study, Princeton, New Jersey 08540 M. H. Kalos Courant Institute of Mathematical Sciences, New York university, New York, New York 10012. Received August 3, 1981 ABSTRACT: We have performed a series of Monte Carlo simulations for a two-dimensional polymer chain with monomers interacting via a Lennard-Jones potential. An analysis of a mean field theory, based on approximating the free energy as the sum of an elastic part and a fluid part, shows that in two dimensions there is a sharp collapse transition, at T = 0, but that there is no ideal or quasi-ideal behavior at the transition as there is in three dimensions. -

Native-Like Mean Structure in the Unfolded Ensemble of Small Proteins
B doi:10.1016/S0022-2836(02)00888-4 available online at http://www.idealibrary.com on w J. Mol. Biol. (2002) 323, 153–164 Native-like Mean Structure in the Unfolded Ensemble of Small Proteins Bojan Zagrovic1, Christopher D. Snow1, Siraj Khaliq2 Michael R. Shirts2 and Vijay S. Pande1,2* 1Biophysics Program The nature of the unfolded state plays a great role in our understanding of Stanford University, Stanford proteins. However, accurately studying the unfolded state with computer CA 94305-5080, USA simulation is difficult, due to its complexity and the great deal of sampling required. Using a supercluster of over 10,000 processors we 2Department of Chemistry have performed close to 800 ms of molecular dynamics simulation in Stanford University, Stanford atomistic detail of the folded and unfolded states of three polypeptides CA 94305-5080, USA from a range of structural classes: the all-alpha villin headpiece molecule, the beta hairpin tryptophan zipper, and a designed alpha-beta zinc finger mimic. A comparison between the folded and the unfolded ensembles reveals that, even though virtually none of the individual members of the unfolded ensemble exhibits native-like features, the mean unfolded structure (averaged over the entire unfolded ensemble) has a native-like geometry. This suggests several novel implications for protein folding and structure prediction as well as new interpretations for experiments which find structure in ensemble-averaged measurements. q 2002 Elsevier Science Ltd. All rights reserved Keywords: mean-structure hypothesis; unfolded state of proteins; *Corresponding author distributed computing; conformational averaging Introduction under folding conditions, with some notable exceptions.14 – 16 This is understandable since under Historically, the unfolded state of proteins has such conditions the unfolded state is an unstable, received significantly less attention than the folded fleeting species making any kind of quantitative state.1 The reasons for this are primarily its struc- experimental measurement very difficult. -
CD of Proteins Sources Include
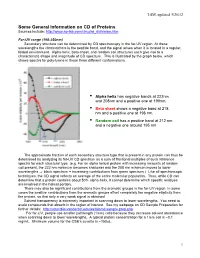
LSM, updated 3/26/12 Some General Information on CD of Proteins Sources include: http://www.ap-lab.com/circular_dichroism.htm Far-UV range (190-250nm) Secondary structure can be determined by CD spectroscopy in the far-UV region. At these wavelengths the chromophore is the peptide bond, and the signal arises when it is located in a regular, folded environment. Alpha-helix, beta-sheet, and random coil structures each give rise to a characteristic shape and magnitude of CD spectrum. This is illustrated by the graph below, which shows spectra for poly-lysine in these three different conformations. • Alpha helix has negative bands at 222nm and 208nm and a positive one at 190nm. • Beta sheet shows a negative band at 218 nm and a positive one at 196 nm. • Random coil has a positive band at 212 nm and a negative one around 195 nm. The approximate fraction of each secondary structure type that is present in any protein can thus be determined by analyzing its far-UV CD spectrum as a sum of fractional multiples of such reference spectra for each structural type. (e.g. For an alpha helical protein with increasing amounts of random coil present, the 222 nm minimum becomes shallower and the 208 nm minimum moves to lower wavelengths ⇒ black spectrum + increasing contributions from green spectrum.) Like all spectroscopic techniques, the CD signal reflects an average of the entire molecular population. Thus, while CD can determine that a protein contains about 50% alpha-helix, it cannot determine which specific residues are involved in the helical portion. -

Large-Scale Analyses of Site-Specific Evolutionary Rates Across
G C A T T A C G G C A T genes Article Large-Scale Analyses of Site-Specific Evolutionary Rates across Eukaryote Proteomes Reveal Confounding Interactions between Intrinsic Disorder, Secondary Structure, and Functional Domains Joseph B. Ahrens, Jordon Rahaman and Jessica Siltberg-Liberles * Department of Biological Sciences, Florida International University, Miami, FL 33199, USA; [email protected] (J.B.A.); jraha001@fiu.edu (J.R.) * Correspondence: jliberle@fiu.edu; Tel.: +1-305-348-7508 Received: 1 October 2018; Accepted: 9 November 2018; Published: 14 November 2018 Abstract: Various structural and functional constraints govern the evolution of protein sequences. As a result, the relative rates of amino acid replacement among sites within a protein can vary significantly. Previous large-scale work on Metazoan (Animal) protein sequence alignments indicated that amino acid replacement rates are partially driven by a complex interaction among three factors: intrinsic disorder propensity; secondary structure; and functional domain involvement. Here, we use sequence-based predictors to evaluate the effects of these factors on site-specific sequence evolutionary rates within four eukaryotic lineages: Metazoans; Plants; Saccharomycete Fungi; and Alveolate Protists. Our results show broad, consistent trends across all four Eukaryote groups. In all four lineages, there is a significant increase in amino acid replacement rates when comparing: (i) disordered vs. ordered sites; (ii) random coil sites vs. sites in secondary structures; and (iii) inter-domain linker sites vs. sites in functional domains. Additionally, within Metazoans, Plants, and Saccharomycetes, there is a strong confounding interaction between intrinsic disorder and secondary structure—alignment sites exhibiting both high disorder propensity and involvement in secondary structures have very low average rates of sequence evolution. -

Protein Folding and Structure Prediction
Protein Folding and Structure Prediction A Statistician's View Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Proteins Amino acids without peptide bonds. Amino acids with peptide bonds. ¡ Amino acids are the building blocks of proteins. Proteins Both figures show the same protein (the bacterial protein L). The right figure also highlights the secondary structure elements. Space Resolution limit of a light microscope Glucose Ribosome Red blood cell C−C bond Hemoglobin Bacterium 1 10 100 1000 10000 100000 1nm 1µm Distance [ A° ] Energy C−C bond Green Noncovalent bond light Glucose Thermal ATP 0.1 1 10 100 1000 Energy [ kcal/mol ] Non-Bonding Interactions Amino acids of a protein are joined by covalent bonding interactions. The polypep- tide is folded in three dimension by non-bonding interactions. These interactions, which can easily be disrupted by extreme pH, temperature, pressure, and denatu- rants, are: Electrostatic Interactions (5 kcal/mol) Hydrogen-bond Interactions (3-7 kcal/mol) Van Der Waals Interactions (1 kcal/mol) ¡ Hydrophobic Interactions ( 10 kcal/mol) The total inter-atomic force acting between two atoms is the sum of all the forces they exert on each other. Energy Profile Transition State Denatured State Native State Radius of Gyration of Denatured Proteins Do chemically denatured proteins behave as random coils? The radius of gyration Rg of a protein is defined as the root mean square dis- tance from each atom of the protein to their centroid. For an ideal (infinitely thin) random-coil chain in a solvent, the average radius 0.5 ¡ of gyration of a random coil is a simple function of its length n: Rg n For an excluded volume polymer (a polymer with non-zero thickness and non- trivial interactions between monomers) in a solvent, the average radius of gyra- 0.588 tion, we have Rg n (Flory 1953). -

Helix-Coil Conformational Change Accompanied by Anisotropic–Isotropic Transition
Polymer Journal, Vol.33, No. 11, pp 898—901 (2001) NOTES Helix-Coil Conformational Change Accompanied by Anisotropic–Isotropic Transition † Ning LIU, Jiaping LIN, Tao C HEN, Jianding CHEN, Dafei ZHOU, and Le LI Department of Polymer Science and Engineering, East China University of Science and Technology, Shanghai 200237, P. R. China (Received November 22, 2000; Accepted July 1, 2001) KEY WORDS Helix-Coil Transition / Liquid Crystal Polymer / Polypeptide / Anisotropic– Isotropic Transition / Polypeptides, particularly poly(γ-benzyl-L- limited study of this acid induced anisotropic–isotropic glutamate) (PBLG) and other glutamic acid ester transition has been reported.13, 14 The precise molecular polymers, have the ability to exist in α-helix, a well de- mechanism of the transition remains to be determined. fined chain conformation of long chain order and retain In the present work, the texture changes in the acid- such a structure in numerous organic solvents that sup- induced anisotropic–isotropic transition were studied port intramolecular hydrogen bonding.1, 2 If PBLG and by microscope observations and the corresponding related polypeptides are dissolved in a binary solvent molecular conformational changes were followed by IR mixture containing a non-helicogenic component such measurements. The effect of the polymer concentra- as dichloroacetic acid (DCA) or trifluoroacetic acid tion was studied. According to the information gained (TFA), they can undergo a conformational change from through the experiment, the molecular mechanism with the α-helix to random coil form due to the changes respect to the acid-induced helix-coil conformational in solvent composition, temperature or both.3, 4 The change was suggested. -

Native-Like Secondary Structure in a Peptide From
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Research Paper 473 Native-like secondary structure in a peptide from the ␣-domain of hen lysozyme Jenny J Yang1*, Bert van den Berg*, Maureen Pitkeathly, Lorna J Smith, Kimberly A Bolin2, Timothy A Keiderling3, Christina Redfield, Christopher M Dobson and Sheena E Radford4 Background: To gain insight into the local and nonlocal interactions that Addresses: Oxford Centre for Molecular Sciences contribute to the stability of hen lysozyme, we have synthesized two peptides and New Chemistry Laboratory, University of Oxford, South Parks Road, Oxford OX1 3QT, UK. that together comprise the entire ␣-domain of the protein. One peptide (peptide Present addresses: 1Department of Molecular 1–40) corresponds to the sequence that forms two ␣-helices, a loop region, and Physics and Biochemistry, Yale University, 266 a small -sheet in the N-terminal region of the native protein. The other (peptide Whitney Avenue, PO Box 208114, New Haven, 84–129) makes up the C-terminal part of the ␣-domain and encompasses two CT 06520-8114, USA. 2Department of Chemistry ␣-helices and a 310 helix in the native protein. and Biochemistry, University of California, Santa Cruz, CA 96064, USA. 3Department of Chemistry, University of Illinois, 845 West Taylor Street, Results: As judged by CD and a range of NMR parameters, peptide 1–40 has Chicago, IL 60607-7061, USA. 4Department of little secondary structure in aqueous solution and only a small number of local Biochemistry and Molecular Biology, University of hydrophobic interactions, largely in the loop region. -

Development of the Semi-Empirical Equation of State for Square-Well Chain Fluid Based on the Statistical Associating Fluid Theory (SAFT)
Korean J. Chem. Eng., 17(1), 52-57 (2000) Development of the Semi-empirical Equation of State for Square-well Chain Fluid Based on the Statistical Associating Fluid Theory (SAFT) Min Sun Yeom, Jaeeon Chang and Hwayong Kim† School of Chemical Engineering, Seoul National University, Shinlim-dong, Kwanak-ku, Seoul 151-742, Korea (Received 25 August 1999 • accepted 29 October 1999) Abstract−A semi-empirical equation of state for the freely jointed square-well chain fluid is developed. This equation of state is based on Wertheim's thermodynamic perturbation theory (TPT) and the statistical associating fluid theory (SAFT). The compressibility factor and radial distribution function of square-well monomer are ob- tained from Monte Carlo simulations. These results are correlated using density expansion. In developing the equa- tion of state the exact analytical expressions are adopted for the second and third virial coefficients for the com- pressibility factor and the first two terms of the radial distribution function, while the higher order coefficients are determined from regression using the simulation data. In the limit of infinite temperature, the present equation of state and the expression for the radial distribution function are represented by the Carnahan-Starling equation of state. This semi-empirical equation of state gives at least comparable accuracy with other empirical equation of state for the square-well monomer fluid. With the new SAFT equation of state from the accurate expressions for the mo- nomer reference and covalent terms, we compare the prediction of the equation of state to the simulation results for the compressibility factor and radial distribution function of the square-well monomer and chain fluids. -

High Elasticity of Polymer Networks. Polymer Networks
High Elasticity of Polymer Networks. Polymer Networks. Polymer network consists of long polymer chains which are crosslinked with each other to form a giant three-dimensional macromolecule. All polymer networks except those who are in the glassy or partially crystalline states exhibit the property of high elasticity, i.e., the ability to undergo large reversible deformations at relatively small applied stress. High elasticity is the most specific property of polymer materials, it is connected with the most fundamental features of ideal chains considered in the previous lecture. In everyday life, highly elastic polymer materials are called rubbers. Molecular Nature of the High Elasticity. Elastic response of the crystalline solids is due to the change of the equilibrium interatomic distances under stress and therefore, the change in the internal energy of the crystal. Elasticity of rubbers is composed from the elastic responses of the chains crosslinked in the network sample. External stress changes the equilibrium end-to-end distance of a chain, and it thus adopts a less probable conformation. Therefore, the elasticity of rubbers is of purely entropic nature. Typical Stress-Strain Curves. for steel for rubbers А - the upper limit for stress-strain linearity, В - the upper limit for the reversibility of deformations, С - the fracture point. - the typical values of deformations ∆ l l are much larger for rubber; - the typical values of strain σ are much larger for steel; - the typical values of the Young modulus is enormously larger for steel than for rubber ( E 1 0 1 1 Pa and 1 0 5 − 1 0 6 Pa, respectively); - for steel linearity and reversibility are lost practically simultaneously, while for rubbers there is a very wide region of nonlinear reversible deformations; - for steel there is a wide region of plastic deformations (between points B and C) which is all but absent for rubbers. -

Non-Covalent Interactions and How Macromolecules Fold
Non-covalent interactions and how macromolecules fold Lecture 6: Protein folding and misfolding Dr Philip Fowler Objective: Restate how proteins fold First-year Biophysics course Examine the thermodynamics of how a protein folds. Determine what factors affect the stability of the folded, native structure. Resolve the Levinthal Paradox Investigate the role misfolded proteins play in disease Summary: Proteins are marginally stable We can study the folding of proteins using e.g. denaturants Proteins can be unfolded by changing pH and temperature or by adding denaturants The concept of the protein folding funnel dispenses with the Levinthal Paradox Misfolded proteins can form aggregates known as fibrils; prions are infectious proteins Protein folding unfolded, extended state compact molten globule folded, compact native state (random coil) more water—water clusters of non-covalent interactions fewer protein—water hydrogen bonds hydrogen bonds (can form form cooperatively complete network) ∆H > 0 ☹ ∆H < 0 ☺ ☺ native fold is less flexible protein than molten globule protein can adopt fewer ∆S < 0 ☹ ∆S < 0 ☹ conformations when a molten globule much less local ordering of the water from the perspective of the ∆H < 0 ☺ ∆H ≈ 0 solvent, there is little difference water between the molten globule and ∆S > 0 ☺ ☺ ∆S ≈ 0 the folded, native state The hydrophobic effect causes the extended polypeptide chain to collapse and form a compact but dynamic molten globule. Clusters of non-covalent interactions within the protein then form cooperatively. Thermodynamics of folding lysozyme at 25 °C DG = DH TDS unfolded, extended state − folded, compact native state ΔG = -60.9 kJ mol-1 TΔS = -175 kJ mol-1 ΔH = -236 kJ mol-1 How much lysozyme is unfolded? includes " includes " (a) protein—protein"☺ (a) increase in entropy of the water "☺ DG = RT lnK (b) protein—water " ☹ (b) decrease in the entropy of the protein ☹ − (c) water—water interactions ☺ K ~ 4.7 x 1010 i.e. -

Introduction to Polymer Physics Lecture 1 Boulder Summer School July – August 3, 2012
Introduction to polymer physics Lecture 1 Boulder Summer School July – August 3, 2012 A.Grosberg New York University Lecture 1: Ideal chains • Course outline: • This lecture outline: •Ideal chains (Grosberg) • Polymers and their •Real chains (Rubinstein) uses • Solutions (Rubinstein) •Scales •Methods (Grosberg) • Architecture • Closely connected: •Polymer size and interactions (Pincus), polyelectrolytes fractality (Rubinstein) , networks • Entropic elasticity (Rabin) , biopolymers •Elasticity at high (Grosberg), semiflexible forces polymers (MacKintosh) • Limits of ideal chain Polymer molecule is a chain: Examples: polyethylene (a), polysterene •Polymericfrom Greek poly- (b), polyvinyl chloride (c)… merēs having many parts; First Known Use: 1866 (Merriam-Webster); • Polymer molecule consists of many elementary units, called monomers; • Monomers – structural units connected by covalent bonds to form polymer; • N number of monomers in a …and DNA polymer, degree of polymerization; •M=N*mmonomer molecular mass. Another view: Scales: •kBT=4.1 pN*nm at • Monomer size b~Å; room temperature • Monomer mass m – (240C) from 14 to ca 1000; • Breaking covalent • Polymerization bond: ~10000 K; degree N~10 to 109; bondsbonds areare NOTNOT inin • Contour length equilibrium.equilibrium. L~10 nm to 1 m. • “Bending” and non- covalent bonds compete with kBT Polymers in materials science (e.g., alkane hydrocarbons –(CH2)-) # C 1-5 6-15 16-25 20-50 1000 or atoms more @ 25oC Gas Low Very Soft solid Tough and 1 viscosity viscous solid atm liquid liquid Uses Gaseous