Complete Sequence and Comparative Analysis of the Chloroplast Genome of Coconut Palm (Cocos Nucifera)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Well-Known Plants in Each Angiosperm Order
Well-known plants in each angiosperm order This list is generally from least evolved (most ancient) to most evolved (most modern). (I’m not sure if this applies for Eudicots; I’m listing them in the same order as APG II.) The first few plants are mostly primitive pond and aquarium plants. Next is Illicium (anise tree) from Austrobaileyales, then the magnoliids (Canellales thru Piperales), then monocots (Acorales through Zingiberales), and finally eudicots (Buxales through Dipsacales). The plants before the eudicots in this list are considered basal angiosperms. This list focuses only on angiosperms and does not look at earlier plants such as mosses, ferns, and conifers. Basal angiosperms – mostly aquatic plants Unplaced in order, placed in Amborellaceae family • Amborella trichopoda – one of the most ancient flowering plants Unplaced in order, placed in Nymphaeaceae family • Water lily • Cabomba (fanwort) • Brasenia (watershield) Ceratophyllales • Hornwort Austrobaileyales • Illicium (anise tree, star anise) Basal angiosperms - magnoliids Canellales • Drimys (winter's bark) • Tasmanian pepper Laurales • Bay laurel • Cinnamon • Avocado • Sassafras • Camphor tree • Calycanthus (sweetshrub, spicebush) • Lindera (spicebush, Benjamin bush) Magnoliales • Custard-apple • Pawpaw • guanábana (soursop) • Sugar-apple or sweetsop • Cherimoya • Magnolia • Tuliptree • Michelia • Nutmeg • Clove Piperales • Black pepper • Kava • Lizard’s tail • Aristolochia (birthwort, pipevine, Dutchman's pipe) • Asarum (wild ginger) Basal angiosperms - monocots Acorales -

ALINTA DBNGP LOOPING 10 Rehabilitation Management Plan
DBNGP (WA) Nominees Pty Ltd DBNGP LOOPING 10 Rehabilitation Management Plan ALINTA DBNGP LOOPING 10 Rehabilitation Management Plan November 2005 Ecos Consulting (Aust) Pty Ltd CONTENTS 1 INTRODUCTION ................................................................................ 1 2 REHABILITATION REVIEW............................................................ 1 2.1 REHABILITATION OBJECTIVES ............................................................... 2 3 EXISTING VEGETATION ................................................................. 2 3.1 FLORA AND VEGETATION...................................................................... 2 3.2 VEGETATION STUDIES ........................................................................... 4 3.2.1 Study Method ............................................................................... 4 3.2.2 Study Results ................................................................................ 7 3.3 OTHER ENVIRONMENTAL VALUES ...................................................... 10 4 REHABILITATION STRATEGY..................................................... 11 5 REHABILITATION METHODS ..................................................... 11 5.1 WEED MANAGEMENT.......................................................................... 11 5.2 DIEBACK (PHYTOPHTHORA CINNAMOMI) MANAGEMENT .................... 11 5.3 PRIORITY AND RARE FLORA MANAGEMENT ........................................ 12 5.4 RESOURCE MANAGEMENT ................................................................... 13 5.5 -

GENOME EVOLUTION in MONOCOTS a Dissertation
GENOME EVOLUTION IN MONOCOTS A Dissertation Presented to The Faculty of the Graduate School At the University of Missouri In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy By Kate L. Hertweck Dr. J. Chris Pires, Dissertation Advisor JULY 2011 The undersigned, appointed by the dean of the Graduate School, have examined the dissertation entitled GENOME EVOLUTION IN MONOCOTS Presented by Kate L. Hertweck A candidate for the degree of Doctor of Philosophy And hereby certify that, in their opinion, it is worthy of acceptance. Dr. J. Chris Pires Dr. Lori Eggert Dr. Candace Galen Dr. Rose‐Marie Muzika ACKNOWLEDGEMENTS I am indebted to many people for their assistance during the course of my graduate education. I would not have derived such a keen understanding of the learning process without the tutelage of Dr. Sandi Abell. Members of the Pires lab provided prolific support in improving lab techniques, computational analysis, greenhouse maintenance, and writing support. Team Monocot, including Dr. Mike Kinney, Dr. Roxi Steele, and Erica Wheeler were particularly helpful, but other lab members working on Brassicaceae (Dr. Zhiyong Xiong, Dr. Maqsood Rehman, Pat Edger, Tatiana Arias, Dustin Mayfield) all provided vital support as well. I am also grateful for the support of a high school student, Cady Anderson, and an undergraduate, Tori Docktor, for their assistance in laboratory procedures. Many people, scientist and otherwise, helped with field collections: Dr. Travis Columbus, Hester Bell, Doug and Judy McGoon, Julie Ketner, Katy Klymus, and William Alexander. Many thanks to Barb Sonderman for taking care of my greenhouse collection of many odd plants brought back from the field. -

Quarterly Changes
Plant Names Database: Quarterly changes 30 November 2015 © Landcare Research New Zealand Limited 2015 This copyright work is licensed under the Creative Commons Attribution 3.0 New Zealand license. Attribution if redistributing to the public without adaptation: "Source: Landcare Research" Attribution if making an adaptation or derivative work: "Sourced from Landcare Research" http://dx.doi.org/doi:10.7931/P1Z598 CATALOGUING IN PUBLICATION Plant names database: quarterly changes [electronic resource]. – [Lincoln, Canterbury, New Zealand] : Landcare Research Manaaki Whenua, 2014- . Online resource Quarterly November 2014- ISSN 2382-2341 I.Manaaki Whenua-Landcare Research New Zealand Ltd. II. Allan Herbarium. Citation and Authorship Wilton, A.D.; Schönberger, I.; Gibb, E.S.; Boardman, K.F.; Breitwieser, I.; Cochrane, M.; Dawson, M.I.; de Pauw, B.; Fife, A.J.; Ford, K.A.; Glenny, D.S.; Heenan, P.B.; Korver, M.A.; Novis, P.M.; Redmond, D.N.; Smissen, R.D. Tawiri, K. (2015) Plant Names Database: Quarterly changes. November 2015. Lincoln, Manaaki Whenua Press. This report is generated using an automated system and is therefore authored by the staff at the Allan Herbarium who currently contribute directly to the development and maintenance of the Plant Names Database. Authors are listed alphabetically after the third author. Authors have contributed as follows: Leadership: Wilton, Heenan, Breitwieser Database editors: Wilton, Schönberger, Gibb Taxonomic and nomenclature research and review: Schönberger, Gibb, Wilton, Breitwieser, Dawson, Ford, Fife, Glenny, Heenan, Novis, Redmond, Smissen Information System development: Wilton, De Pauw, Cochrane Technical support: Boardman, Korver, Redmond, Tawiri Disclaimer The Plant Names Database is being updated every working day. We welcome suggestions for improvements, concerns, or any data errors you may find. -

Vascular Plants of Santa Cruz County, California
ANNOTATED CHECKLIST of the VASCULAR PLANTS of SANTA CRUZ COUNTY, CALIFORNIA SECOND EDITION Dylan Neubauer Artwork by Tim Hyland & Maps by Ben Pease CALIFORNIA NATIVE PLANT SOCIETY, SANTA CRUZ COUNTY CHAPTER Copyright © 2013 by Dylan Neubauer All rights reserved. No part of this publication may be reproduced without written permission from the author. Design & Production by Dylan Neubauer Artwork by Tim Hyland Maps by Ben Pease, Pease Press Cartography (peasepress.com) Cover photos (Eschscholzia californica & Big Willow Gulch, Swanton) by Dylan Neubauer California Native Plant Society Santa Cruz County Chapter P.O. Box 1622 Santa Cruz, CA 95061 To order, please go to www.cruzcps.org For other correspondence, write to Dylan Neubauer [email protected] ISBN: 978-0-615-85493-9 Printed on recycled paper by Community Printers, Santa Cruz, CA For Tim Forsell, who appreciates the tiny ones ... Nobody sees a flower, really— it is so small— we haven’t time, and to see takes time, like to have a friend takes time. —GEORGIA O’KEEFFE CONTENTS ~ u Acknowledgments / 1 u Santa Cruz County Map / 2–3 u Introduction / 4 u Checklist Conventions / 8 u Floristic Regions Map / 12 u Checklist Format, Checklist Symbols, & Region Codes / 13 u Checklist Lycophytes / 14 Ferns / 14 Gymnosperms / 15 Nymphaeales / 16 Magnoliids / 16 Ceratophyllales / 16 Eudicots / 16 Monocots / 61 u Appendices 1. Listed Taxa / 76 2. Endemic Taxa / 78 3. Taxa Extirpated in County / 79 4. Taxa Not Currently Recognized / 80 5. Undescribed Taxa / 82 6. Most Invasive Non-native Taxa / 83 7. Rejected Taxa / 84 8. Notes / 86 u References / 152 u Index to Families & Genera / 154 u Floristic Regions Map with USGS Quad Overlay / 166 “True science teaches, above all, to doubt and be ignorant.” —MIGUEL DE UNAMUNO 1 ~ACKNOWLEDGMENTS ~ ANY THANKS TO THE GENEROUS DONORS without whom this publication would not M have been possible—and to the numerous individuals, organizations, insti- tutions, and agencies that so willingly gave of their time and expertise. -

Perth and Peel Green Growth Plan for 3.5 Million
v Government of Western Australia Department of the Premier and Cabinet Perth and Peel Green Growth Plan for 3.5 million Strategic Assessment of the Perth and Peel Regions Draft EPBC Act Strategic Impact Assessment Report Part D: MNES Assessment$IBQUFS December 2015 Acknowledgements This document has been prepared by Eco Logical Australia Pty Ltd and © Government of Western Australia Open Lines Consulting. Department of the Premier and Cabinet Disclaimer Dumas House This document may only be used for the purpose for which it was 2 Havelock Street commissioned and in accordance with the contract between Eco Logical Australia Pty Ltd and the Department of the Premier and West Perth WA 6005 Cabinet. The scope of services was defined in consultation with the Department of the Premier and Cabinet, by time and budgetary Website: www.dpc.wa.gov.au/greengrowthplan constraints imposed by the client, and the availability of reports Email: [email protected] and other data on the subject area. Changes to available information, legislation and schedules are made on an ongoing Tel: 08 6552 5151 basis and readers should obtain up to date information. Fax: 08 6552 5001 Eco Logical Australia Pty Ltd accepts no liability or responsibility Published December 2015 whatsoever for or in respect of any use of or reliance upon this report and its supporting material by any third party. Information provided is not intended to be a substitute for site specific assessment or legal advice in relation to any matter. Unauthorised use of this report in any form is prohibited. Strategic As sessment for the Perth and Peel Regions Contents List of Figures.......................................................................................................................................................... -

On the Flora of Australia
L'IBRARY'OF THE GRAY HERBARIUM HARVARD UNIVERSITY. BOUGHT. THE FLORA OF AUSTRALIA, ITS ORIGIN, AFFINITIES, AND DISTRIBUTION; BEING AN TO THE FLORA OF TASMANIA. BY JOSEPH DALTON HOOKER, M.D., F.R.S., L.S., & G.S.; LATE BOTANIST TO THE ANTARCTIC EXPEDITION. LONDON : LOVELL REEVE, HENRIETTA STREET, COVENT GARDEN. r^/f'ORElGN&ENGLISH' <^ . 1859. i^\BOOKSELLERS^.- PR 2G 1.912 Gray Herbarium Harvard University ON THE FLORA OF AUSTRALIA ITS ORIGIN, AFFINITIES, AND DISTRIBUTION. I I / ON THE FLORA OF AUSTRALIA, ITS ORIGIN, AFFINITIES, AND DISTRIBUTION; BEIKG AN TO THE FLORA OF TASMANIA. BY JOSEPH DALTON HOOKER, M.D., F.R.S., L.S., & G.S.; LATE BOTANIST TO THE ANTARCTIC EXPEDITION. Reprinted from the JJotany of the Antarctic Expedition, Part III., Flora of Tasmania, Vol. I. LONDON : LOVELL REEVE, HENRIETTA STREET, COVENT GARDEN. 1859. PRINTED BY JOHN EDWARD TAYLOR, LITTLE QUEEN STREET, LINCOLN'S INN FIELDS. CONTENTS OF THE INTRODUCTORY ESSAY. § i. Preliminary Remarks. PAGE Sources of Information, published and unpublished, materials, collections, etc i Object of arranging them to discuss the Origin, Peculiarities, and Distribution of the Vegetation of Australia, and to regard them in relation to the views of Darwin and others, on the Creation of Species .... iii^ § 2. On the General Phenomena of Variation in the Vegetable Kingdom. All plants more or less variable ; rate, extent, and nature of variability ; differences of amount and degree in different natural groups of plants v Parallelism of features of variability in different groups of individuals (varieties, species, genera, etc.), and in wild and cultivated plants vii Variation a centrifugal force ; the tendency in the progeny of varieties being to depart further from their original types, not to revert to them viii Effects of cross-impregnation and hybridization ultimately favourable to permanence of specific character x Darwin's Theory of Natural Selection ; — its effects on variable organisms under varying conditions is to give a temporary stability to races, species, genera, etc xi § 3. -

Flora Recovery Plan: Threatened Tasmanian Grasstrees 2006-2010
DEPARTMENT OF PRIMARY INDUSTRIES AND WATER Threatened Tasmanian Grasstrees Xanthorrhoea arenaria and Xanthorrhoea bracteata Flora Recovery Plan 2006 - 2010 Threatened Tasmanian Grasstrees Flora Recovery Plan 2006-2010 1 DISCLAIMER This recovery plan recognises that identification of Xanthorrhoea to species level can be difficult as plants often exhibit intermediate or combined characters of the listed species, Xanthorrhoea bracteata and Xanthorrhoea arenaria and the non-listed Xanthorrhoea australis. The identification difficulty arises from the large degree of variation inherent within each of these species and considerable overlap of the variation between species as well as reports of hybridisation and introgression between species. The identity of populations which have closer affinities to either Xanthorrhoea arenaria or Xanthorrhoea bracteata have therefore often been recorded by observers as Xanthorrhoea aff. arenaria or Xanthorrhoea aff. bracteata respectively, though this rarely, if ever, precludes the existence of individual plants within those populations that could be ascribed to either Xanthorrhoea arenaria or Xanthorrhoea bracteata. In a few cases, observers could not be contacted to confirm the occurrence of either Xanthorrhoea arenaria or Xanthorrhoea bracteata at such sites and they are included in the population table in this plan as they were recorded (Xanthorrhoea aff. bracteata). Until the occurrence of Xanthorrhoea arenaria or Xanthorrhoea bracteata can be verified at these sites, it is recommended that such populations be considered as threatened and treated accordingly. ACKNOWLEDGEMENTS This plan was prepared by Eve Lazarus (TSS) with advice from Wendy Potts (TSS), Tim Rudman (Vegetation Section, BCB), Alex Buchanan and Dennis Morris (Tasmanian Herbarium), Richard Barnes (Private Forest Reserves Program, DPIW), Richard Schahinger, Mark Wapstra (Forest Practices Authority) and Tim Wardlaw (Forestry Tasmania), Alison Woolley (Forestry Tasmania/ Conservation Assessment Section, DPIW) and Steve Casey (Conservation Assessment Section, DPIW). -
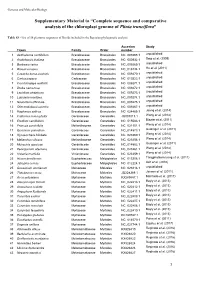
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Plinia Trunciflora”
Genetics and Molecular Biology Supplementary Material to “Complete sequence and comparative analysis of the chloroplast genome of Plinia trunciflora” Table S3 - List of 56 plastome sequences of Rosids included in the Bayesian phylogenetic analysis. Accesion Study Taxon Family Order number 1 Aethionema cordifolium Brassicaceae Brassicales NC_009265.1 unpublished 2 Arabidopsis thaliana Brassicaceae Brassicales NC_000932.1 Sato et al. (1999) 3 Barbarea verna Brassicaceae Brassicales NC_009269.1 unpublished 4 Brassica napus Brassicaceae Brassicales NC_016734.1 Hu et al. (2011) 5 Capsella bursa-pastoris Brassicaceae Brassicales NC_009270.1 unpublished 6 Carica papaya Caricaceae Brassicales NC_010323.1 unpublished 7 Crucihimalaya wallichii Brassicaceae Brassicales NC_009271.1 unpublished 8 Draba nemorosa Brassicaceae Brassicales NC_009272.1 unpublished 9 Lepidium virginicum Brassicaceae Brassicales NC_009273.1 unpublished 10 Lobularia maritima Brassicaceae Brassicales NC_009274.1 unpublished 11 Nasturtium officinale Brassicaceae Brassicales NC_009275.1 unpublished 12 Olimarabidopsis pumila Brassicaceae Brassicales NC_009267.1 unpublished 13 Raphanus sativus Brassicaceae Brassicales NC_024469.1 Jeong et al. (2014) 14 California macrophylla Geraniaceae Geraniales JQ031013.1 Weng et al. (2014) 15 Erodium carvifolium Geraniaceae Geraniales NC_015083.1 Blazier et al. (2011) 16 Francoa sonchifolia Melianthaceae Geraniales NC_021101.1 Weng et al. (2014) 17 Geranium palmatum Geraniaceae Geraniales NC_014573.1 Guisinger et al. (2011) 18 Hypseocharis bilobate Geraniaceae Geraniales NC_023260.1 Weng et al. (2014) 19 Melianthus villosus Melianthaceae Geraniales NC_023256.1 Weng et al. (2014) 20 Monsonia speciose Geraniaceae Geraniales NC_014582.1 Guisinger et al. (2011) 21 Pelargonium alternans Geraniaceae Geraniales NC_023261.1 Weng et al. (2014) 22 Viviania marifolia Vivianiaceae Geraniales NC_023259.1 Weng et al. (2014) 23 Hevea brasiliensis Euphorbiaceae Malpighiales NC_015308.1 Tangphatsornruang et al. -

Downloaded from Genbank on That Full Plastid Genomes Are Not Sufficient to Reject Al- February 28, 2012
Ruhfel et al. BMC Evolutionary Biology 2014, 14:23 http://www.biomedcentral.com/1471-2148/14/23 RESEARCH ARTICLE Open Access From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes Brad R Ruhfel1*, Matthew A Gitzendanner2,3,4, Pamela S Soltis3,4, Douglas E Soltis2,3,4 and J Gordon Burleigh2,4 Abstract Background: Next-generation sequencing has provided a wealth of plastid genome sequence data from an increasingly diverse set of green plants (Viridiplantae). Although these data have helped resolve the phylogeny of numerous clades (e.g., green algae, angiosperms, and gymnosperms), their utility for inferring relationships across all green plants is uncertain. Viridiplantae originated 700-1500 million years ago and may comprise as many as 500,000 species. This clade represents a major source of photosynthetic carbon and contains an immense diversity of life forms, including some of the smallest and largest eukaryotes. Here we explore the limits and challenges of inferring a comprehensive green plant phylogeny from available complete or nearly complete plastid genome sequence data. Results: We assembled protein-coding sequence data for 78 genes from 360 diverse green plant taxa with complete or nearly complete plastid genome sequences available from GenBank. Phylogenetic analyses of the plastid data recovered well-supported backbone relationships and strong support for relationships that were not observed in previous analyses of major subclades within Viridiplantae. However, there also is evidence of systematic error in some analyses. In several instances we obtained strongly supported but conflicting topologies from analyses of nucleotides versus amino acid characters, and the considerable variation in GC content among lineages and within single genomes affected the phylogenetic placement of several taxa. -

Flora of Australia, Volume 46, Iridaceae to Dioscoreaceae
FLORA OF AUSTRALIA Volume 46 Iridaceae to Dioscoreaceae This volume was published before the Commonwealth Government moved to Creative Commons Licensing. © Commonwealth of Australia 1986. This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced or distributed by any process or stored in any retrieval system or data base without prior written permission from the copyright holder. Requests and inquiries concerning reproduction and rights should be addressed to: [email protected] FLORA OF AUSTRALIA The nine families in this volume of the Flora of Australia are Iridaceae, Aloeaceae, Agavaceae, Xanthorrhoeaceae, Hanguan- aceae, Taccaceae, Stemonaceae, Smilacaceae and Dioscoreaceae. The Xanthorrhoeaceae has the largest representation with 10 genera and 99 species. Most are endemic with a few species of Lomandra and Romnalda extending to neighbouring islands. The family includes the spectacular blackboys and grass-trees. The Iridaceae is largely represented by naturalised species with 52 of the 78 species being introduced. Many of the introductions are ornamentals and several have become serious weeds. Patersonia is the largest genus with all 17 species endemic. Some of these are cultivated as ornamentals. The Dioscoreaccae is a family of economic significance, particularly in the old world tropics where some species are cultivated or collected for their tubers and bulbils. In Australia there are 5 species, one of which is a recent introduction. The endemic and native species, commonly known as yams, are traditionally eaten by the Aborigines. -

Checklist of the Vascular Plants of San Diego County 5Th Edition
cHeckliSt of tHe vaScUlaR PlaNtS of SaN DieGo coUNty 5th edition Pinus torreyana subsp. torreyana Downingia concolor var. brevior Thermopsis californica var. semota Pogogyne abramsii Hulsea californica Cylindropuntia fosbergii Dudleya brevifolia Chorizanthe orcuttiana Astragalus deanei by Jon P. Rebman and Michael G. Simpson San Diego Natural History Museum and San Diego State University examples of checklist taxa: SPecieS SPecieS iNfRaSPecieS iNfRaSPecieS NaMe aUtHoR RaNk & NaMe aUtHoR Eriodictyon trichocalyx A. Heller var. lanatum (Brand) Jepson {SD 135251} [E. t. subsp. l. (Brand) Munz] Hairy yerba Santa SyNoNyM SyMBol foR NoN-NATIVE, NATURaliZeD PlaNt *Erodium cicutarium (L.) Aiton {SD 122398} red-Stem Filaree/StorkSbill HeRBaRiUM SPeciMeN coMMoN DocUMeNTATION NaMe SyMBol foR PlaNt Not liSteD iN THE JEPSON MANUAL †Rhus aromatica Aiton var. simplicifolia (Greene) Conquist {SD 118139} Single-leaF SkunkbruSH SyMBol foR StRict eNDeMic TO SaN DieGo coUNty §§Dudleya brevifolia (Moran) Moran {SD 130030} SHort-leaF dudleya [D. blochmaniae (Eastw.) Moran subsp. brevifolia Moran] 1B.1 S1.1 G2t1 ce SyMBol foR NeaR eNDeMic TO SaN DieGo coUNty §Nolina interrata Gentry {SD 79876} deHeSa nolina 1B.1 S2 G2 ce eNviRoNMeNTAL liStiNG SyMBol foR MiSiDeNtifieD PlaNt, Not occURRiNG iN coUNty (Note: this symbol used in appendix 1 only.) ?Cirsium brevistylum Cronq. indian tHiStle i checklist of the vascular plants of san Diego county 5th edition by Jon p. rebman and Michael g. simpson san Diego natural history Museum and san Diego state university publication of: san Diego natural history Museum san Diego, california ii Copyright © 2014 by Jon P. Rebman and Michael G. Simpson Fifth edition 2014. isBn 0-918969-08-5 Copyright © 2006 by Jon P.