Ex Situ Collections and Their Potential for the Restoration of Extinct Plants
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Investigation of Plant Species with Identified Seed Oil Fatty Acids In
ORIGINAL RESEARCH published: 22 February 2017 doi: 10.3389/fpls.2017.00224 Investigation of Plant Species with Identified Seed Oil Fatty Acids in Chinese Literature and Analysis of Five Unsurveyed Chinese Endemic Species Changsheng Li 1, Xiaojun Cheng 1, Qingli Jia 1, Huan Song 1, Xiangling Liu 1, Kai Wang 1, Cuizhu Zhao 1, Yansheng Zhang 2, John Ohlrogge 3 and Meng Zhang 1* 1 Plant Science Department, College of Agronomy, Northwest A&F University, Yangling, China, 2 CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, Edited by: China, 3 Department of Plant Biology, Michigan State University, East Lansing, MI, USA Basil J. Nikolau, Iowa State University, USA Reviewed by: Diverse fatty acid structures from different plant species are important renewable Xiao Qiu, resources for industrial raw materials and as liquid fuels with high energy density. Because University of Saskatchewan, Canada of its immense geographical and topographical variations, China is a country with Yonghua Li-Beisson, The French Atomic Energy and enormous diversity of plant species, including large numbers of plants endemic to China. Alternative Energies Commission The richness of this resource of species provides a wide range of fatty acids in seeds or (CEA), France other tissues, many of which have been identified by Chinese scientists. However, in *Correspondence: Meng Zhang the past, most publications describing analysis of these plants were written in Chinese, [email protected] making access for researchers from other countries difficult. In this study, we investigated reports on seed and fruit oil fatty acids as described in Chinese literature. -
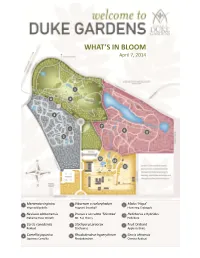
What's in Bloom
WHAT’S IN BLOOM April 7, 2014 5 4 6 2 7 1 9 8 3 12 10 11 1 Mertensia virginica 5 Viburnum x carlcephalum 9 Malus ‘Hopa’ Virginia Bluebells Fragrant Snowball Flowering Crabapple 2 Neviusia alabamensis 6 Prunus x serrulata ‘Shirotae’ 10 Helleborus x hybridus Alabama Snow Wreath Mt. Fuji Cherry Hellebore 3 Cercis canadensis 7 Stachyurus praecox 11 Fruit Orchard Redbud Stachyurus Apple cultivars 4 Camellia japonica 8 Rhododendron hyperythrum 12 Cercis chinensis Japanese Camellia Rhododendron Chinese Redbud WHAT’S IN BLOOM April 7, 2014 BLOMQUIST GARDEN OF NATIVE PLANTS Amelanchier arborea Common Serviceberry Sanguinaria canadensis Bloodroot Cornus florida Flowering Dogwood Stylophorum diphyllum Celandine Poppy Thalictrum thalictroides Rue Anemone Fothergilla major Fothergilla Trillium decipiens Chattahoochee River Trillium Hepatica nobilis Hepatica Trillium grandiflorum White Trillium Hexastylis virginica Wild Ginger Hexastylis minor Wild Ginger Trillium pusillum Dwarf Wakerobin Illicium floridanum Florida Anise Tree Trillium stamineum Blue Ridge Wakerobin Malus coronaria Sweet Crabapple Uvularia sessilifolia Sessileleaf Bellwort Mertensia virginica Virginia Bluebells Pachysandra procumbens Allegheny spurge Prunus americana American Plum DORIS DUKE CENTER GARDENS Camellia japonica Japanese Camellia Pulmonaria ‘Diana Clare’ Lungwort Cercis canadensis Redbud Prunus persica Flowering Peach Puschkinia scilloides Striped Squill Cercis chinensis Redbud Sanguinaria canadensis Bloodroot Clematis armandii Evergreen Clematis Spiraea prunifolia Bridalwreath -

Comparative Genomics of the Balsaminaceae Sister Genera Hydrocera Triflora and Impatiens Pinfanensis
International Journal of Molecular Sciences Article Comparative Genomics of the Balsaminaceae Sister Genera Hydrocera triflora and Impatiens pinfanensis Zhi-Zhong Li 1,2,†, Josphat K. Saina 1,2,3,†, Andrew W. Gichira 1,2,3, Cornelius M. Kyalo 1,2,3, Qing-Feng Wang 1,3,* and Jin-Ming Chen 1,3,* ID 1 Key Laboratory of Aquatic Botany and Watershed Ecology, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan 430074, China; [email protected] (Z.-Z.L.); [email protected] (J.K.S.); [email protected] (A.W.G.); [email protected] (C.M.K.) 2 University of Chinese Academy of Sciences, Beijing 100049, China 3 Sino-African Joint Research Center, Chinese Academy of Sciences, Wuhan 430074, China * Correspondence: [email protected] (Q.-F.W.); [email protected] (J.-M.C.); Tel.: +86-27-8751-0526 (Q.-F.W.); +86-27-8761-7212 (J.-M.C.) † These authors contributed equally to this work. Received: 21 December 2017; Accepted: 15 January 2018; Published: 22 January 2018 Abstract: The family Balsaminaceae, which consists of the economically important genus Impatiens and the monotypic genus Hydrocera, lacks a reported or published complete chloroplast genome sequence. Therefore, chloroplast genome sequences of the two sister genera are significant to give insight into the phylogenetic position and understanding the evolution of the Balsaminaceae family among the Ericales. In this study, complete chloroplast (cp) genomes of Impatiens pinfanensis and Hydrocera triflora were characterized and assembled using a high-throughput sequencing method. The complete cp genomes were found to possess the typical quadripartite structure of land plants chloroplast genomes with double-stranded molecules of 154,189 bp (Impatiens pinfanensis) and 152,238 bp (Hydrocera triflora) in length. -

Plastome Structure and Phylogenetic Relationships of Styracaceae (Ericales)
Plastome Structure and Phylogenetic Relationships of Styracaceae (Ericales) Xiu-lian Cai Hainan University Jacob B. Landis Cornell University Hong-Xin Wang Hainan University Jian-Hua Wang Hainan University Zhi-Xin Zhu Hainan University Huafeng Wang ( [email protected] ) Hainan University https://orcid.org/0000-0002-0218-1728 Review Keywords: Styracaceae, Plastome, Genome structure, Phylogeny, positive selection Posted Date: January 29th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-55283/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/25 Abstract Background: The Styracaceae are a woody, dicotyledonous family containing 12 genera and an estimated 160 species. Recent studies have shown that Styrax and Sinojackia are monophyletic, Alniphyllum and Bruinsmia cluster into a clade with an approximately 20-kb inversion in the Large Single-Copy (LSC) region. Halesia and Pterostyrax are not supported as monophyletic, while Melliodendron and Changiostyrax always form sister clades . Perkinsiodendron and Changiostyrax were newly established genera of Styracaceae. However, the phylogenetic relationship of Styracaceae at the genera level needs further research. Results: We collected 28 complete plastomes of Styracaceae, including 12 sequences newly reported here and 16 publicly available complete plastome sequences, comprising 11 of the 12 genera of Styracaceae. All species possessed the typical quadripartite structure of angiosperm plastomes, and the sequence difference is small, except for the large 20-kb (14 genes) inversion region found in Alniphyllum and Bruinsmia. Seven coding sequences (rps4, rpl23, accD, rpoC1, psaA, rpoA and ndhH) were identied to possess positively selected sites. Phylogenetic reconstructions based on seven data sets (i.e., LSC, SSC, IR, Coding, Non-coding, combination of LSC+SSC and concatenation of LSC+SSC+one IR) produced similar topologies. -

Seven Complete Chloroplast Genomes from Symplocos: Genome Organization and Comparative Analysis
Article Seven Complete Chloroplast Genomes from Symplocos: Genome Organization and Comparative Analysis Sang-Chul Kim 1 , Jei-Wan Lee 1,* and Byoung-Ki Choi 2 1 Department of Forest Bioresources, National Institute of Forest Science, Suwon 16631, Korea; [email protected] 2 Warm Temperate and Subtropical Forest Research Center, National Institute of Forest Science, 22, Donnaeko-Ro, Seogwipo-Si 63582, Korea; [email protected] * Correspondence: [email protected] Abstract: In the present study, chloroplast genome sequences of four species of Symplocos (S. chinensis for. pilosa, S. prunifolia, S. coreana, and S. tanakana) from South Korea were obtained by Ion Torrent sequencing and compared with the sequences of three previously reported Symplocos chloroplast genomes from different species. The length of the Symplocos chloroplast genome ranged from 156,961 to 157,365 bp. Overall, 132 genes including 87 functional genes, 37 tRNA genes, and eight rRNA genes were identified in all Symplocos chloroplast genomes. The gene order and contents were highly similar across the seven species. The coding regions were more conserved than the non- coding regions, and the large single-copy and small single-copy regions were less conserved than the inverted repeat regions. We identified five new hotspot regions (rbcL, ycf4, psaJ, rpl22, and ycf1) that can be used as barcodes or species-specific Symplocos molecular markers. These four novel chloroplast genomes provide basic information on the plastid genome of Symplocos and enable better taxonomic characterization of this genus. Citation: Kim, S.-C.; Lee, J.-W.; Choi, B.-K. Seven Complete Chloroplast Keywords: chloroplast; genome; next-generation sequencing; phylogenetics; simple sequence repeat Genomes from Symplocos: Genome Organization and Comparative Analysis. -

China: a Rich Flora Needed of Urgent Conservationprovided by Digital.CSIC
Orsis 19, 2004 49-89 View metadata, citation and similar papers at core.ac.uk brought to you by CORE China: a rich flora needed of urgent conservationprovided by Digital.CSIC López-Pujol, Jordi GReB, Laboratori de Botànica, Facultat de Farmàcia, Universitat de Barcelona, Avda. Joan XXIII s/n, E-08028, Barcelona, Catalonia, Spain. Author for correspondence (E-mail: [email protected]) Zhao, A-Man Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, The People’s Republic of China. Manuscript received in april 2004 Abstract China is one of the richest countries in plant biodiversity in the world. Besides to a rich flora, which contains about 33 000 vascular plants (being 30 000 of these angiosperms, 250 gymnosperms, and 2 600 pteridophytes), there is a extraordinary ecosystem diversity. In addition, China also contains a large pool of both wild and cultivated germplasm; one of the eight original centers of crop plants in the world was located there. China is also con- sidered one of the main centers of origin and diversification for seed plants on Earth, and it is specially profuse in phylogenetically primitive taxa and/or paleoendemics due to the glaciation refuge role played by this area in the Quaternary. The collision of Indian sub- continent enriched significantly the Chinese flora and produced the formation of many neoen- demisms. However, the distribution of the flora is uneven, and some local floristic hotspots can be found across China, such as Yunnan, Sichuan and Taiwan. Unfortunately, threats to this biodiversity are huge and have increased substantially in the last 50 years. -

Styracaceae R.Br
Styracaceae R.Br. Alniphyllum Matsum. Halesia J.Ellis ex L. Huodendron Rehder Melliodendron Hand.-Mazz. Pterostyrax Siebold & Zucc. Rehderodendron Hu Sinojackia Hu Styrax L. VEGETATIVE KEY TO SPECIES IN CULTIVATION Jan De Langhe (23 October 2012 - 18 January 2016) Vegetative identification key. Introduction: This key is based on vegetative characteristics, and therefore also of use when flowers and fruits are absent. - Use a 10× hand lens to check pubescence, bud scales, teeth and venation pattern in general. - Start counting veins at base of the top leaves lamina with first clearly ascending secondary vein, do not include intercalary veins, nor these ending in the apex. - Look at the entire plant. Avoid young specimens, shade- and strong shoots as these give an atypical view. - Beware of hybridisation, especially with plants raised from seed other than wild origin. Taxa treated in this key: see page 6. Taxa referred to synonymy in this key: see page 6. Questionable/frequently misapplied names: see page 6. References: - JDL herbarium - living specimens, in various arboreta, botanic gardens and collections - literature: Bean, W.J. & Clarke, D.L. - (1981-1988) - Styracaceae in Bean's Trees and Shrubs hardy in the British Isles - and online edition Grimshaw, J. & Bayton, R. - (2009) - Styracaceae in New Trees, 976p. Huang, S. & Grimes J.W. - (1996) - Styracaceae in Flora of China VOL.15, p.253-271 - and online edition Huang, Y, Fritsch, P.W. & Shi, S. - (2003) - A revision of the imbricate group of Styrax series Cyrta in Asia - Ann. Missouri Bot. Gard. VOL. 90:p.491-553. Krüssmann, G. - (1977-1978) - Styracaceae in Handbuch der Laubgehölze, 3 VOL. -

Great Plants for Southern Gardens That Missed the Marketing Push©
Great Plants for Southern Gardens That Missed the Marketing Push 447 Great Plants for Southern Gardens That Missed the Marketing Push© Mark Weathington JC Raulston Arboretum at North Carolina State University, Department of Horticultural Sci- ence, Campus Box 7522, Raleigh, North Carolina 27695 Email: [email protected] INTRODUCTION The JC Raulston Arboretum (JCRA) has a 35-year history of collecting and evalu- ating new plants for introduction to the nursery industry. New plants drive the industry especially those that have a marketing push behind them. Many great landscape plants get passed over in the rush for the latest and greatest but deserve a second look. The JC Raulston Arboretum (JCRA) has grown from the first plant the JCRA planted in the mid-1970s. Conifers which were not supposed to survive in the south have grown into mature specimens. Gardens and collections have been planted, grown up, torn out, and re-established. Students have been the mainstay of the arboretum development and have done a great job when given adequate direction. The JCRA’s collection holds many great landscape plants that may never make it to the mainstream due to propagation difficulties. Other plants have shown they have great potential for the south. DECIDUOUS SHRUBS Rhododendron ‘Yoshino’ has proven its worth in the arboretum with leathery, semi- evergreen foliage and trusses of bright flowers in March. The growth habit is much different than many other evergreen azaleas and is used extensively as a hedging plant in Japan. Hydrangea hirta is an underused member of the genus that suf- fers from not having the sterile florets of other landscape hydrangeas. -

A Consensus Phylogenomic Approach Highlights Paleopolyploid and Rapid Radiation in the History of Ericales
RESEARCH ARTICLE A consensus phylogenomic approach highlights paleopolyploid and rapid radiation in the history of Ericales Drew A. Larson1,4 , Joseph F. Walker2 , Oscar M. Vargas3 , and Stephen A. Smith1 Manuscript received 8 December 2019; revision accepted 12 February PREMISE: Large genomic data sets offer the promise of resolving historically recalcitrant 2020. species relationships. However, different methodologies can yield conflicting results, 1 Department of Ecology & Evolutionary Biology, University of especially when clades have experienced ancient, rapid diversification. Here, we analyzed Michigan, Ann Arbor, MI 48109, USA the ancient radiation of Ericales and explored sources of uncertainty related to species tree 2 Sainsbury Laboratory (SLCU), University of Cambridge, Cambridge, inference, conflicting gene tree signal, and the inferred placement of gene and genome CB2 1LR, UK duplications. 3 Department of Ecology & Evolutionary Biology, University of California, Santa Cruz, CA 95060, USA METHODS: We used a hierarchical clustering approach, with tree-based homology and 4Author for correspondence (e-mail: [email protected]) orthology detection, to generate six filtered phylogenomic matrices consisting of data Citation: Larson, D. A., J. F. Walker, O. M. Vargas, and S. A. Smith. from 97 transcriptomes and genomes. Support for species relationships was inferred 2020. A consensus phylogenomic approach highlights paleopolyploid from multiple lines of evidence including shared gene duplications, gene tree conflict, and rapid radiation -

WUCOLS List S Abelia Chinensis Chinese Abelia M ? ? M / / Copyright © UC Regents, Davis Campus
Ba Bu G Gc P Pm S Su T V N Botanical Name Common Name 1 2 3 4 5 6 Symbol Vegetation Used in Type WUCOLS List S Abelia chinensis Chinese abelia M ? ? M / / Copyright © UC Regents, Davis campus. All rights reserved. bamboo Ba S Abelia floribunda Mexican abelia M ? M M / / S Abelia mosanensis 'Fragrant Abelia' fragrant abelia ? ? ? ? ? ? bulb Bu S Abelia parvifolia (A. longituba) Schuman abelia ? ? ? M ? ? grass G groundcover GC Gc S Abelia x grandiflora and cvs. glossy abelia M M M M M / perennial* P S Abeliophyllum distichum forsythia M M ? ? ? ? palm and cycad Pm S Abelmoschus manihot (Hibiscus manihot) sunset muskmallow ? ? ? L ? ? T Abies pinsapo Spanish fir L L L / / / shrub S succulent Su T N Abies spp. (CA native and non-native) fir M M M M / / P N Abronia latifolia yellow sand verbena VL VL VL / ? ? tree T P N Abronia maritima sand verbena VL VL VL / ? ? vine V California N native S N Abutilon palmeri Indian mallow L L L L M M S Abutilon pictum thompsonii variegated Chinese lantern M H M M ? ? Sunset WUCOLS CIMIS ET Representative Number climate 0 Region zones** Cities zones* S Abutilon vitifolium flowering maple M M M / ? ? Healdsburg, Napa, North- San Jose, Salinas, Central 14, 15, 16, 17 1, 2, 3, 4, 6, 8 San Francisco, Coastal San Luis Obispo S Abutilon x hybridum & cvs. flowering maple M H M M / / 1 Auburn, Central Bakersfield, Chico, 8, 9, 14 12, 14, 15, 16 Valley Fresno, Modesto, Sacramento S T Acacia abyssinica Abyssinian acacia / ? / ? / L 2 Irvine, Los South Angeles, Santa 22, 23, 24 1, 2, 4, 6 Coastal Barbara, Ventura, -
Styracaceae R.Br
Styracaceae R.Br. Alniphyllum Matsum. Halesia J.Ellis ex L. Huodendron Rehder Melliodendron Hand.-Mazz. Perkinsiodendron P.W.Fritsch Pterostyrax Siebold & Zucc. Rehderodendron Hu Sinojackia Hu Styrax L. VEGETATIVE KEY TO SPECIES IN CULTIVATION Jan De Langhe (23 October 2012 - 27 February 2019) Vegetative identification key. Introduction: This key is based on vegetative characteristics, and therefore also of use when flowers and fruits are absent. - Use a 10× hand lens to check pubescence, bud scales, teeth and venation pattern in general. - Start counting veins at base of the top leaves lamina with first clearly ascending secondary vein, do not include intercalary veins, nor these ending in the apex. - Look at the entire plant. Avoid young specimens, shade- and strong shoots as these give an atypical view. - Beware of hybridisation, especially with plants raised from seed other than wild origin. Taxa treated in this key: see page 6. Taxa referred to synonymy in this key: see page 6. Questionable/frequently misapplied names: see page 6. References: - JDL herbarium - living specimens, in various arboreta, botanic gardens and collections - literature: Bean, W.J. & Clarke, D.L. - (1981-1988) - Styracaceae in Bean's Trees and Shrubs hardy in the British Isles - and online edition Grimshaw, J. & Bayton, R. - (2009) - Styracaceae in New Trees, 976p. Huang, S. & Grimes J.W. - (1996) - Styracaceae in Flora of China VOL.15, p.253-271 - and online edition Huang, Y, Fritsch, P.W. & Shi, S. - (2003) - A revision of the imbricate group of Styrax series Cyrta in Asia - Ann. Missouri Bot. Gard. VOL. 90:p.491-553. Krüssmann, G. -

List of Replacement Tree Species for Planting Trees on Private Property
Urban Forestry 1900 SW 4th Ave. Suite 5000 Portland, OR 97201 Email: [email protected] Tel: (503) 823-TREE (8733) Fax: (503) 823-4493 List of Replacement Tree Species For planting trees on private property Sustaining a healthy park and recreation system to make Portland a great place to live, work and play. www.PortlandOregon.gov/trees • Commissioner Amanda Fritz • Director Mike Abbaté Replace removed trees with something of value Trees vary greatly in how well they cool and clean the air, slow rainfall runoff, quiet sound, and serve as habitat. When choosing trees to replace those being removed, consider species that provide the highest value to the urban environment: - Evergreens: provide year-round benefits - Natives: adapted to Portland’s climate - Large trees: provide more canopy benefits than small trees - Disease-resistant species or cultivars: improve the urban forest’s resilience Also consider trees that: - Are longer-lived - Thrive in Portland’s climate - Are drought tolerant, reducing water demand in the summer - Provide habitat for wildlife (flowers for pollinators or seeds, fruits, or cones for birds and other wildlife) Minimum requirements for a replacement tree Replacement trees must: • Grow on average to at least 15 feet or more • Be reliably hardy in the Portland area • Not be an invasive species (nuisance tree) on the Portland Plant List • Not be prone to fatal pests or diseases, such as Dutch elm disease How to reduce the number of replacement trees you are required to plant Select the species you wish to plant from the following lists. You can reduce the number of trees you are required to plant by choosing to plant evergreen trees.