Seven Complete Chloroplast Genomes from Symplocos: Genome Organization and Comparative Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mobile Genetic Elements in Streptococci
Curr. Issues Mol. Biol. (2019) 32: 123-166. DOI: https://dx.doi.org/10.21775/cimb.032.123 Mobile Genetic Elements in Streptococci Miao Lu#, Tao Gong#, Anqi Zhang, Boyu Tang, Jiamin Chen, Zhong Zhang, Yuqing Li*, Xuedong Zhou* State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, PR China. #Miao Lu and Tao Gong contributed equally to this work. *Address correspondence to: [email protected], [email protected] Abstract Streptococci are a group of Gram-positive bacteria belonging to the family Streptococcaceae, which are responsible of multiple diseases. Some of these species can cause invasive infection that may result in life-threatening illness. Moreover, antibiotic-resistant bacteria are considerably increasing, thus imposing a global consideration. One of the main causes of this resistance is the horizontal gene transfer (HGT), associated to gene transfer agents including transposons, integrons, plasmids and bacteriophages. These agents, which are called mobile genetic elements (MGEs), encode proteins able to mediate DNA movements. This review briefly describes MGEs in streptococci, focusing on their structure and properties related to HGT and antibiotic resistance. caister.com/cimb 123 Curr. Issues Mol. Biol. (2019) Vol. 32 Mobile Genetic Elements Lu et al Introduction Streptococci are a group of Gram-positive bacteria widely distributed across human and animals. Unlike the Staphylococcus species, streptococci are catalase negative and are subclassified into the three subspecies alpha, beta and gamma according to the partial, complete or absent hemolysis induced, respectively. The beta hemolytic streptococci species are further classified by the cell wall carbohydrate composition (Lancefield, 1933) and according to human diseases in Lancefield groups A, B, C and G. -

The Right Tree Trail: Species Selection Information
The Right Tree Trail: Species selection information All the below species are available through TreeSearch Farms in Houston, and are hardy to our area. These species will grow to an appropriate height which will not conflict with the safety of overhead utilities. Cercis canadensis ‘Texensis’ Texas Redbud Native, 15’ x 12’, heart-shaped semi-glossy foliage, magnificent display of rosy-pink flowers in spring, sun/part shade, requires good drainage, deciduous Clethera pringlei Clethera Evergreen, 20’, upright growth, dark green glossy foliage, racemes of fragrant creamy white flowers in summer with the aroma of cinnamon, full sun/light shade Sinojackia rehderiana Sinojackia 20’, fast growing, sculptured trunk, semi-glossy foliage, clusters of white star-shaped flowers in spring, sun/part shade, deciduous Vitex agnus-castus ‘Montrose Purple’ Montrose Purple Vitex 18’ multi-trunk, new vitex with slightly larger foliage & magnificent clusters of 8”-12” bloom spikes 3 times larger than the standard vitex with richer, dark blue flowers, blooms at least 3 times spring to fall, sun, drought tolerant, butterflies Ilex vomitoria ‘Pumphouse Red’ Pumphouse Red Yaupon Evergreen, 18’-18’, large shrub/small tree, bluish-green foliage, produces hundreds of red berries, full sun/part shade, tolerant of soil, water, & lighting Viburnum rufidulum Lord Byron Viburnum 15’, slow growing, small tree, glossy dark, evergreen foliage, blooms white 4”-6” clusters in spring, foliage turns red, mauve, purple, orange & yellow in fall, fruit ripens in fall, full sun/part -

Symplocos Paniculata
CHAPTER P14 PLEASE USE ONLY US ENGLISH; you must follow the format strictly; Browse through this sample chapter before typing yours in US ENGLISH only SYMPLOCOS PANICULATA (SAPPHIRE BERRY): A WOODY AND ENERGY- EFFICIENT OIL PLANT QIANG LIU, YOUPING SUN, JINGZHENG CHEN, PEIWANG LI, GENHUA NIU, CHANGZHU LI AND LIJUAN JIANG all names = first, middle & last Qiang Liu, PhD Candidate, Central South University of Forestry and Technology, 498 South Shaoshan Rd., Changsha, Hunan 410004, China. Mobile: 86-15116272702; E-mail: [email protected] we must have mailing address/email of all authors where ecopy of book will be sent Youping Sun [first name, middle initial, last name], PhD, Senior Research Associate, Texas A&M AgriLife Research Center at El Paso, 1380 A&M Circle, El Paso, TX 79927, USA. Mobile: +1-8646339385; E-mail: [email protected] Indicate Job title for all authors Jingzheng Chen, PhD Candidate, Research Scientist, Hunan Academy of Forestry, 658 South Shaoshan Rd., Changsha, Hunan 410004, China. Mobile: +86-15111084183; E-mail: [email protected] Peiwang Li, M.S., Associate Professor, Hunan Academy of Forestry, 658 South Shaoshan Rd., Changsha, Hunan 410004, China. Mobile: +86-15116325230; E-mail: [email protected] Genhua Niu, PhD, Associate Professor, Texas A&M AgriLife Research Center at El Paso, 1380 A&M Circle, El Paso, TX 79927, USA. Mobile: +1-9152082993; E-mail: [email protected] Lijuan Jiang, PhD, Professor, Central South University of Forestry and Technology, 498 South Shaoshan Rd., Changsha, Hunan 410004, China. Mobile: +86-13407314259; E-mail: [email protected] Changzhu Li, Ph.D., Professor, Hunan Academy of Forestry, 658 South Shaoshan Rd., Changsha, Hunan 410004, China. -

Ethno Botanical Polypharmacy of Traditional Healers in Wayanad (Kerala) to Treat Type 2 Diabetes
Indian Journal of Traditional Knowledge Vol. 11(4), October 2012, pp. 667-673 Ethno Botanical Polypharmacy of Traditional Healers in Wayanad (Kerala) to treat type 2 diabetes Dilip Kumar EK & Janardhana GR* Phytopharmacology Laboratory, Department of Studies in Botany University of Mysore, Manasagangothri, Mysore-570006, Karnataka, India E-mail: [email protected] Received 30.06.10, revised 15.05.12 The aboriginal medical system prevalent among traditional healers of Wayanad has demonstrated a good practice, so bright future in the therapy of type 2 diabetes. Therefore, present study focused on identification validation and documentation such Ethno botanical polypharmacy prevalent in the district. A total of 47 species belonging to 44 genera comes under 29 families were identified being utilized in 23 different compound medicinal recipes for diabetic healthcare in Wayanad. These preparations and the herbal ingredients need scientific evaluation about their mechanism of action in living organism in heath as well as disease condition to confirm their activity against type 2 diabetes. Keywords: Type 2 diabetes, Traditional medicine, Polypharmacy, Wayanad district IPC Int. Cl.8: A61K, A61K 36/00, A01D 16/02, A01D 16/03 Local herbal healers of Wayanad (Kerala), India have communities that directly depend on it. The present numerous prescriptions aims directly to treat and study documented some of the ethno botanical manage type 2 diabetes (old age diabetes). This remedies for the management of diabetes so as includes over 150 herbal preparations including to protect it within the aboriginal repository of simple and compound folk recipes and diets. This knowledge (ARK) programme and also shed light traditional medical knowledge has demonstrated a on a traditional culture that believes that a healthy potent therapeutic system for the management of lifestyle is found only at a healthy environment 1. -

Approaches and Limitations of Species Level Diagnostics in Flowering Plants
Genetic Food Diagnostics Approaches and Limitations of Species Level Diagnostics in Flowering Plants Zur Erlangung des akademischen Grades eines DOKTORS DER NATURWISSENSCHAFTEN (Dr. rer. nat.) Fakultät für Chemie und Biowissenschaften Karlsruher Institut für Technologie (KIT) - Universitätsbereich genehmigte DISSERTATION von Dipl. Biologe Thomas Horn aus 77709 Wolfach Dekan: Prof. Dr. Peter Roesky Referent: Prof. Dr. Peter Nick Korreferent: Prof. Dr. Horst Taraschewski Tag der mündlichen Prüfung: 17.04.2014 Parts of this work are derived from the following publications: Horn T, Völker J, Rühle M, Häser A, Jürges G, Nick P; 2013; Genetic authentication by RFLP versus ARMS? The case of Moldavian Dragonhead (Dracocephalum moldavica L.). European Food Research and Technology, doi 10.1007/s00217-013-2089-4 Horn T, Barth A, Rühle M, Häser A, Jürges G, Nick P; 2012; Molecular Diagnostics of Lemon Myrtle (Backhousia citriodora versus Leptospermum citratum). European Food Research and Technology, doi 10.1007/s00217-012-1688-9 Also included are works from the following teaching projects: RAPD Analysis and SCAR design in the TCM complex Clematis Armandii Caulis (chuān mù tōng), F2 Plant Evolution, 2011 Effects of highly fragmented DNA on PCR, F3, Lidija Krebs, 2012 1 I. Acknowledgement “Nothing is permanent except change” Heraclitus of Ephesus Entering adolescence – approximately 24 years ago – many aspects of life pretty much escaped my understanding. After a period of turmoil and subsequent experience of a life as laborer lacking an education, I realized that I did not want to settle for this kind of life. I wanted to change. With this work I would like to thank all people that ever bothered trying to explain the world to me, that allowed me to find my way and nurtured my desire to change. -

Wood Anatomy of Korean Symplocos Jacq. (Symplocaceae)
Korean J. Pl. Taxon. 50(3): 333−342 (2020) pISSN 1225-8318 eISSN 2466-1546 https://doi.org/10.11110/kjpt.2020.50.3.333 Korean Journal of RESEARCH ARTICLE Plant Taxonomy Wood anatomy of Korean Symplocos Jacq. (Symplocaceae) Balkrishna GHIMIRE, Beom Kyun PARK, Seung-Hwan OH, Jaedong LEE1 and Dong Chan SON* Division of Forest Biodiversity, Korea National Arboretum, Pocheon 11186, Korea 1Division of Biological Sciences, Jeonbuk National University, Jeonju 54896, Korea (Received 25 June 2020; Revised 16 September 2020; Accepted 23 September 2020) ABSTRACT: Despite poorly documented species delimitation and unresolved taxonomic nomenclature, four spe- cies of Symplocos (S. coreana, S purnifolia, S sawafutagi, and S. tanakana) have been described in Korea. In this study, we carried a comparative wood anatomy analysis of all four species of Korean Symplocos to understand the wood anatomical variations among them. The results of this study indicated that Korean Symplocos are com- paratively indistinguishable in terms of their qualitative wood features, except for exclusively uniseriate rays present in S. purnifolia instead of uniseriate to multiseriate in other three species. Nevertheless, differences are noticed in quantitative wood variables such as the vessel density, vessel size, and ray density. The vessel density of S. purnifolia is more than twice as high as those of S. sawafutagi and S. tanakana. In contrast, the vessel circumference and diameter on both plants of S. sawafutagi and S. tanakana is nearly twice as large as those of S. purnifolia. Sym- plocos coreana has characteristic intermediacy between these two groups in terms of vessel features and is closer to S. -

From the Human Genome Project to Genomic Medicine a Journey to Advance Human Health
From the Human Genome Project to Genomic Medicine A Journey to Advance Human Health Eric Green, M.D., Ph.D. Director, NHGRI The Origin of “Genomics”: 1987 Genomics (1987) “For the newly developing discipline of [genome] mapping/sequencing (including the analysis of the information), we have adopted the term GENOMICS… ‘The Genome Institute’ Office for Human Genome Research 1988-1989 National Center for Human Genome Research 1989-1997 National Human Genome Research Institute 1997-present NHGRI: Circa 1990-2003 Human Genome Project NHGRI Today: Characteristic Features . Relatively young (~28 years) . Relatively small (~1.7% of NIH) . Unusual historical origins (think ‘Human Genome Project’) . Emphasis on ‘Team Science’ (think managed ‘consortia’) . Rapidly disseminating footprint (think ‘genomics’) . Novel societal/bioethics research component (think ‘ELSI’) . Over-achievers for trans-NIH initiatives (think ‘Common Fund’) . Vibrant (and large) Intramural Research Program A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project Genomic Medicine An emerging medical discipline that involves using genomic information about an individual as part of their clinical care (e.g., for diagnostic or therapeutic decision- making) and the other implications of that clinical use The Path to Genomic Medicine ? Human Realization of Genome Genomic Project Medicine Nature Nature Base Pairs to Bedside 2003 Heli201x to 1Health A Quarter Century of Genomics Human Genome Sequenced for First Time by the Human Genome Project -

Halesia Spp. Family: Styracaceae Silverbell
Halesia spp. Family: Styracaceae Silverbell The genus Halesia is composed of about 4 species native to: the United States [3] and China [1]. The word halesia is named after Stephen Hales (1677-1761), British clergyman and author of Vegetable Staticks (1722). Halesia carolina-Bell-tree, Bell Olivetree, Bellwood, Box-elder, Carolina Silverbell, Catbell, Florida Silverbell, Four-winged Halesia, Little Silverbell, No-name-tree, Opossum, Opossumwood, Mountain Silverbell, Rattle-box, Silverbell-tree, Silver-tree, Snowdrop-tree, Tisswood, Wild Olivetree Halesia diptera-Cowlicks, Silverbell-tree, Snowdrop-tree, Southern Silverbell-tree, Two Wing Silverbell Halesia parviflora-Florida Silverbell, Little Silverbell. Distribution Southeastern United States and China. The Tree Silverbells are shrubs or trees with scaly reddish brown bark. The leaves and small branches are covered with stellate (star shaped) hairs. The showy white flowers are produced in small, pendulous clusters. They produce dry, winged fruits (samara). Silverbells can reach a height of 100 feet, although they normally grow to 40 feet. The bark is thin, separating into slightly ridged, reddish brown scales. The Wood General The wood of Silverbell is brown, strong, dense and close grained. It has a wide white sapwood and a pale brown heartwood. The luster is medium and it has no odor or taste. The texture is fine and uniform, with a straight grain. Mechanical Properties (2-inch standard) Compression Specific MOE MOR Parallel Perpendicular WMLa Hardness Shear gravity GPa MPa MPa MPa kJ/m3 N MPa Green .42 8.0 11.8 19.5 3.0 60.7 2090 6.4 Dry .48 9.1 59.3 35.4 4.7 47.6 2624 8.1 aWML = Work to maximum load. -

Investigation of Plant Species with Identified Seed Oil Fatty Acids In
ORIGINAL RESEARCH published: 22 February 2017 doi: 10.3389/fpls.2017.00224 Investigation of Plant Species with Identified Seed Oil Fatty Acids in Chinese Literature and Analysis of Five Unsurveyed Chinese Endemic Species Changsheng Li 1, Xiaojun Cheng 1, Qingli Jia 1, Huan Song 1, Xiangling Liu 1, Kai Wang 1, Cuizhu Zhao 1, Yansheng Zhang 2, John Ohlrogge 3 and Meng Zhang 1* 1 Plant Science Department, College of Agronomy, Northwest A&F University, Yangling, China, 2 CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, Edited by: China, 3 Department of Plant Biology, Michigan State University, East Lansing, MI, USA Basil J. Nikolau, Iowa State University, USA Reviewed by: Diverse fatty acid structures from different plant species are important renewable Xiao Qiu, resources for industrial raw materials and as liquid fuels with high energy density. Because University of Saskatchewan, Canada of its immense geographical and topographical variations, China is a country with Yonghua Li-Beisson, The French Atomic Energy and enormous diversity of plant species, including large numbers of plants endemic to China. Alternative Energies Commission The richness of this resource of species provides a wide range of fatty acids in seeds or (CEA), France other tissues, many of which have been identified by Chinese scientists. However, in *Correspondence: Meng Zhang the past, most publications describing analysis of these plants were written in Chinese, [email protected] making access for researchers from other countries difficult. In this study, we investigated reports on seed and fruit oil fatty acids as described in Chinese literature. -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Book of Abstracts.Pdf
1 List of presenters A A., Hudson 329 Anil Kumar, Nadesa 189 Panicker A., Kingman 329 Arnautova, Elena 150 Abeli, Thomas 168 Aronson, James 197, 326 Abu Taleb, Tariq 215 ARSLA N, Kadir 363 351Abunnasr, 288 Arvanitis, Pantelis 114 Yaser Agnello, Gaia 268 Aspetakis, Ioannis 114 Aguilar, Rudy 105 Astafieff, Katia 80, 207 Ait Babahmad, 351 Avancini, Ricardo 320 Rachid Al Issaey , 235 Awas, Tesfaye 354, 176 Ghudaina Albrecht , Matthew 326 Ay, Nurhan 78 Allan, Eric 222 Aydınkal, Rasim 31 Murat Allenstein, Pamela 38 Ayenew, Ashenafi 337 Amat De León 233 Azevedo, Carine 204 Arce, Elena An, Miao 286 B B., Von Arx 365 Bétrisey, Sébastien 113 Bang, Miin 160 Birkinshaw, Chris 326 Barblishvili, Tinatin 336 Bizard, Léa 168 Barham, Ellie 179 Bjureke, Kristina 186 Barker, Katharine 220 Blackmore, 325 Stephen Barreiro, Graciela 287 Blanchflower, Paul 94 Barreiro, Graciela 139 Boillat, Cyril 119, 279 Barteau, Benjamin 131 Bonnet, François 67 Bar-Yoseph, Adi 230 Boom, Brian 262, 141 Bauters, Kenneth 118 Boratyński, Adam 113 Bavcon, Jože 111, 110 Bouman, Roderick 15 Beck, Sarah 217 Bouteleau, Serge 287, 139 Beech, Emily 128 Bray, Laurent 350 Beech, Emily 135 Breman, Elinor 168, 170, 280 Bellefroid, Elke 166, 118, 165 Brockington, 342 Samuel Bellet Serrano, 233, 259 Brockington, 341 María Samuel Berg, Christian 168 Burkart, Michael 81 6th Global Botanic Gardens Congress, 26-30 June 2017, Geneva, Switzerland 2 C C., Sousa 329 Chen, Xiaoya 261 Cable, Stuart 312 Cheng, Hyo Cheng 160 Cabral-Oliveira, 204 Cho, YC 49 Joana Callicrate, Taylor 105 Choi, Go Eun 202 Calonje, Michael 105 Christe, Camille 113 Cao, Zhikun 270 Clark, John 105, 251 Carta, Angelino 170 Coddington, 220 Carta Jonathan Caruso, Emily 351 Cole, Chris 24 Casimiro, Pedro 244 Cook, Alexandra 212 Casino, Ana 276, 277, 318 Coombes, Allen 147 Castro, Sílvia 204 Corlett, Richard 86 Catoni, Rosangela 335 Corona Callejas , 274 Norma Edith Cavender, Nicole 84, 139 Correia, Filipe 204 Ceron Carpio , 274 Costa, João 244 Amparo B. -
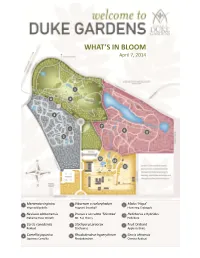
What's in Bloom
WHAT’S IN BLOOM April 7, 2014 5 4 6 2 7 1 9 8 3 12 10 11 1 Mertensia virginica 5 Viburnum x carlcephalum 9 Malus ‘Hopa’ Virginia Bluebells Fragrant Snowball Flowering Crabapple 2 Neviusia alabamensis 6 Prunus x serrulata ‘Shirotae’ 10 Helleborus x hybridus Alabama Snow Wreath Mt. Fuji Cherry Hellebore 3 Cercis canadensis 7 Stachyurus praecox 11 Fruit Orchard Redbud Stachyurus Apple cultivars 4 Camellia japonica 8 Rhododendron hyperythrum 12 Cercis chinensis Japanese Camellia Rhododendron Chinese Redbud WHAT’S IN BLOOM April 7, 2014 BLOMQUIST GARDEN OF NATIVE PLANTS Amelanchier arborea Common Serviceberry Sanguinaria canadensis Bloodroot Cornus florida Flowering Dogwood Stylophorum diphyllum Celandine Poppy Thalictrum thalictroides Rue Anemone Fothergilla major Fothergilla Trillium decipiens Chattahoochee River Trillium Hepatica nobilis Hepatica Trillium grandiflorum White Trillium Hexastylis virginica Wild Ginger Hexastylis minor Wild Ginger Trillium pusillum Dwarf Wakerobin Illicium floridanum Florida Anise Tree Trillium stamineum Blue Ridge Wakerobin Malus coronaria Sweet Crabapple Uvularia sessilifolia Sessileleaf Bellwort Mertensia virginica Virginia Bluebells Pachysandra procumbens Allegheny spurge Prunus americana American Plum DORIS DUKE CENTER GARDENS Camellia japonica Japanese Camellia Pulmonaria ‘Diana Clare’ Lungwort Cercis canadensis Redbud Prunus persica Flowering Peach Puschkinia scilloides Striped Squill Cercis chinensis Redbud Sanguinaria canadensis Bloodroot Clematis armandii Evergreen Clematis Spiraea prunifolia Bridalwreath