Ptprot-Mediated Regulation of P53&Sol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Methylation and Silencing of Protein Tyrosine Phosphatase Receptor
Human Cancer Biology Methylation and Silencing of Protein Tyrosine Phosphatase Receptor Type O in Chronic Lymphocytic Leukemia Tasneem Motiwala,1Sarmila Majumder,1Huban Kutay,1David Spencer Smith,1Donna S. Neuberg,4 David M. Lucas,2 John C. Byrd,2,3 Michael Grever,2,3 and Samson T.Jacob1, 2 , 3 Abstract Purpose: Previous studies in our laboratory have shown the progressive methylation and suppression of the gene encoding protein tyrosine phosphatase, PTPRO, in the livers of rats fed a methyl-deficient diet that induces hepatocarcinogenesis. Subsequently, we observed the methylation of PTPRO in primary human lung tumors and also showed its potential tumor suppressor characteristics. The present study was undertaken to investigate whether the truncated form of PTPRO (PTPROt), specifically expressed in naI«ve B lymphocytes, was also methylated and suppressed in chronic lymphocytic leukemia (CLL), a disease generally affecting B lymphocytes. Experimental Design and Results: Initial screening showed that 60% of the 52 CLL samples analyzed using methylation-specific PCR assay were methylated compared with B lymphocytes from normal individuals, which were not methylated. The expression of PTPROt, as measured by semiquantitative reverse transcription-PCR, inversely correlated with methylation in the few samples tested. Analysis of additional samples (n = 50) by combined bisulfite restriction analysis showed that the PTPRO CpG island was methylated in 82% of patients with CLL compared with B lymphocytes from normal individuals. Furthermore, overall expression of PTPRO was reduced in CLL relative to normal lymphocytes. The PTPRO gene was also suppressed by methylation in the CLL cell lineWaC3CD5, where it could be reactivated upon treatment with the DNA hypome- thylating agent 5-AzaC. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Regulatory Roles of Phosphatases in Cancer
Oncogene (2014) 33, 939–953 & 2014 Macmillan Publishers Limited All rights reserved 0950-9232/14 www.nature.com/onc REVIEW The regulatory roles of phosphatases in cancer J Stebbing1, LC Lit1, H Zhang, RS Darrington, O Melaiu, B Rudraraju and G Giamas The relevance of potentially reversible post-translational modifications required for controlling cellular processes in cancer is one of the most thriving arenas of cellular and molecular biology. Any alteration in the balanced equilibrium between kinases and phosphatases may result in development and progression of various diseases, including different types of cancer, though phosphatases are relatively under-studied. Loss of phosphatases such as PTEN (phosphatase and tensin homologue deleted on chromosome 10), a known tumour suppressor, across tumour types lends credence to the development of phosphatidylinositol 3--kinase inhibitors alongside the use of phosphatase expression as a biomarker, though phase 3 trial data are lacking. In this review, we give an updated report on phosphatase dysregulation linked to organ-specific malignancies. Oncogene (2014) 33, 939–953; doi:10.1038/onc.2013.80; published online 18 March 2013 Keywords: cancer; phosphatases; solid tumours GASTROINTESTINAL MALIGNANCIES abs in sera were significantly associated with poor survival in Oesophageal cancer advanced ESCC, suggesting that they may have a clinical utility in Loss of PTEN (phosphatase and tensin homologue deleted on ESCC screening and diagnosis.5 chromosome 10) expression in oesophageal cancer is frequent, Cao et al.6 investigated the role of protein tyrosine phosphatase, among other gene alterations characterizing this disease. Zhou non-receptor type 12 (PTPN12) in ESCC and showed that PTPN12 et al.1 found that overexpression of PTEN suppresses growth and protein expression is higher in normal para-cancerous tissues than induces apoptosis in oesophageal cancer cell lines, through in 20 ESCC tissues. -

Targeting Protein Tyrosine Phosphatases in Cancer Lakshmi Reddy Bollu, Abhijit Mazumdar, Michelle I
Published OnlineFirst January 13, 2017; DOI: 10.1158/1078-0432.CCR-16-0934 Molecular Pathways Clinical Cancer Research Molecular Pathways: Targeting Protein Tyrosine Phosphatases in Cancer Lakshmi Reddy Bollu, Abhijit Mazumdar, Michelle I. Savage, and Powel H. Brown Abstract The aberrant activation of oncogenic signaling pathways is a act as tumor suppressor genes by terminating signal responses universal phenomenon in cancer and drives tumorigenesis and through the dephosphorylation of oncogenic kinases. More malignant transformation. This abnormal activation of signal- recently, it has become clear that several PTPs overexpressed ing pathways in cancer is due to the altered expression of in human cancers do not suppress tumor growth; instead, they protein kinases and phosphatases. In response to extracellular positively regulate signaling pathways and promote tumor signals, protein kinases activate downstream signaling path- development and progression. In this review, we discuss both ways through a series of protein phosphorylation events, ulti- types of PTPs: those that have tumor suppressor activities as mately producing a signal response. Protein tyrosine phospha- well as those that act as oncogenes. We also discuss the tases (PTP) are a family of enzymes that hydrolytically remove potential of PTP inhibitors for cancer therapy. Clin Cancer Res; phosphate groups from proteins. Initially, PTPs were shown to 23(9); 1–7. Ó2017 AACR. Background in cancer and discuss the current status of PTP inhibitors for cancer therapy. Signal transduction is a complex process that transmits extra- PTPs belong to a superfamily of enzymes that hydrolytically cellular signals effectively through a cascade of events involving remove phosphate groups from proteins (2). -

Application of Microrna Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease
Article Application of microRNA Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease Jennifer L. Major, Rushita A. Bagchi * and Julie Pires da Silva * Department of Medicine, Division of Cardiology, University of Colorado Anschutz Medical Campus, Aurora, CO 80045, USA; [email protected] * Correspondence: [email protected] (R.A.B.); [email protected] (J.P.d.S.) Supplementary Tables Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 www.mdpi.com/journal/mps Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 2 of 25 Table 1. List of all hsa-miRs identified by Human microRNA Disease Database (HMDD; v3.2) analysis. hsa-miRs were identified using the term “genetics” and “circulating” as input in HMDD. Targets CAD hsa-miR-1 Targets IR injury hsa-miR-423 Targets Obesity hsa-miR-499 hsa-miR-146a Circulating Obesity Genetics CAD hsa-miR-423 hsa-miR-146a Circulating CAD hsa-miR-149 hsa-miR-499 Circulating IR Injury hsa-miR-146a Circulating Obesity hsa-miR-122 Genetics Stroke Circulating CAD hsa-miR-122 Circulating Stroke hsa-miR-122 Genetics Obesity Circulating Stroke hsa-miR-26b hsa-miR-17 hsa-miR-223 Targets CAD hsa-miR-340 hsa-miR-34a hsa-miR-92a hsa-miR-126 Circulating Obesity Targets IR injury hsa-miR-21 hsa-miR-423 hsa-miR-126 hsa-miR-143 Targets Obesity hsa-miR-21 hsa-miR-223 hsa-miR-34a hsa-miR-17 Targets CAD hsa-miR-223 hsa-miR-92a hsa-miR-126 Targets IR injury hsa-miR-155 hsa-miR-21 Circulating CAD hsa-miR-126 hsa-miR-145 hsa-miR-21 Targets Obesity hsa-mir-223 hsa-mir-499 hsa-mir-574 Targets IR injury hsa-mir-21 Circulating IR injury Targets Obesity hsa-mir-21 Targets CAD hsa-mir-22 hsa-mir-133a Targets IR injury hsa-mir-155 hsa-mir-21 Circulating Stroke hsa-mir-145 hsa-mir-146b Targets Obesity hsa-mir-21 hsa-mir-29b Methods Protoc. -

Mycoplasma Mycoides Subsp
RESEARCH ARTICLE Whole Blood Transcriptome Analysis of Mycoplasma mycoides Subsp. mycoides-Infected Cattle Confirms Immunosuppression but Does Not Reflect Local Inflammation Valérie Rodrigues1,2*, Philippe Holzmuller1,2, Carinne Puech1,2, Hezron Wesonga3, François Thiaucourt1,2, Lucía Manso-Silván1,2 1 CIRAD, UMR15 CMAEE, F-34398 Montpellier, France, 2 INRA, UMR1309 CMAEE, F-34398 Montpellier, France, 3 Kenyan Agricultural Research Institute, Nairobi, Kenya * [email protected] OPEN ACCESS Abstract Citation: Rodrigues V, Holzmuller P, Puech C, Wesonga H, Thiaucourt F, Manso-Silván L (2015) Contagious bovine pleuropneumonia (CBPP), caused by Mycoplasma mycoides subsp. Mycoplasma Whole Blood Transcriptome Analysis of mycoides (Mmm), is a severe respiratory disease of cattle responsible for major economic mycoides Subsp. mycoides-Infected Cattle Confirms Immunosuppression but Does Not Reflect Local losses in sub-Saharan Africa. Disease control relies mainly on the use of empirically attenu- Inflammation. PLoS ONE 10(10): e0139678. ated vaccines that provide limited protection. Thus, understanding the virulence mechanisms doi:10.1371/journal.pone.0139678 used by Mmm as well as the role of the host immune system in disease development, persis- Editor: Bernhard Kaltenboeck, Auburn University, tence, and control is a prerequisite for the development of new, rationally designed control UNITED STATES strategies. The aim of this study was to assess the use of whole blood transcriptome analysis Received: February 12, 2015 to study cattle-Mmm interactions, starting by the characterization of the bovine response to Accepted: September 16, 2015 Mmm infection during the acute form of the disease. For that purpose, we compared the tran- scriptome profile of whole blood from six cattle, before challenge by contact with Mmm- Published: October 2, 2015 infected animals and at the appearance of first clinical signs, using a bovine microarray. -

Phosphatases Page 1
Phosphatases esiRNA ID Gene Name Gene Description Ensembl ID HU-05948-1 ACP1 acid phosphatase 1, soluble ENSG00000143727 HU-01870-1 ACP2 acid phosphatase 2, lysosomal ENSG00000134575 HU-05292-1 ACP5 acid phosphatase 5, tartrate resistant ENSG00000102575 HU-02655-1 ACP6 acid phosphatase 6, lysophosphatidic ENSG00000162836 HU-13465-1 ACPL2 acid phosphatase-like 2 ENSG00000155893 HU-06716-1 ACPP acid phosphatase, prostate ENSG00000014257 HU-15218-1 ACPT acid phosphatase, testicular ENSG00000142513 HU-09496-1 ACYP1 acylphosphatase 1, erythrocyte (common) type ENSG00000119640 HU-04746-1 ALPL alkaline phosphatase, liver ENSG00000162551 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPPL2 alkaline phosphatase, placental-like 2 ENSG00000163286 HU-07767-1 BPGM 2,3-bisphosphoglycerate mutase ENSG00000172331 HU-06476-1 BPNT1 3'(2'), 5'-bisphosphate nucleotidase 1 ENSG00000162813 HU-09086-1 CANT1 calcium activated nucleotidase 1 ENSG00000171302 HU-03115-1 CCDC155 coiled-coil domain containing 155 ENSG00000161609 HU-09022-1 CDC14A CDC14 cell division cycle 14 homolog A (S. cerevisiae) ENSG00000079335 HU-11533-1 CDC14B CDC14 cell division cycle 14 homolog B (S. cerevisiae) ENSG00000081377 HU-06323-1 CDC25A cell division cycle 25 homolog A (S. pombe) ENSG00000164045 HU-07288-1 CDC25B cell division cycle 25 homolog B (S. pombe) ENSG00000101224 HU-06033-1 CDKN3 cyclin-dependent kinase inhibitor 3 ENSG00000100526 HU-02274-1 CTDSP1 CTD (carboxy-terminal domain, -

Molecular Pathways: Targeting Protein Tyrosine Phosphatases in Cancer Lakshmi Reddy Bollu, Abhijit Mazumdar, Michelle I
Published OnlineFirst January 13, 2017; DOI: 10.1158/1078-0432.CCR-16-0934 Molecular Pathways Clinical Cancer Research Molecular Pathways: Targeting Protein Tyrosine Phosphatases in Cancer Lakshmi Reddy Bollu, Abhijit Mazumdar, Michelle I. Savage, and Powel H. Brown Abstract The aberrant activation of oncogenic signaling pathways is a act as tumor suppressor genes by terminating signal responses universal phenomenon in cancer and drives tumorigenesis and through the dephosphorylation of oncogenic kinases. More malignant transformation. This abnormal activation of signal- recently, it has become clear that several PTPs overexpressed ing pathways in cancer is due to the altered expression of in human cancers do not suppress tumor growth; instead, they protein kinases and phosphatases. In response to extracellular positively regulate signaling pathways and promote tumor signals, protein kinases activate downstream signaling path- development and progression. In this review, we discuss both ways through a series of protein phosphorylation events, ulti- types of PTPs: those that have tumor suppressor activities as mately producing a signal response. Protein tyrosine phospha- well as those that act as oncogenes. We also discuss the tases (PTP) are a family of enzymes that hydrolytically remove potential of PTP inhibitors for cancer therapy. Clin Cancer Res; phosphate groups from proteins. Initially, PTPs were shown to 23(9); 1–7. Ó2017 AACR. Background in cancer and discuss the current status of PTP inhibitors for cancer therapy. Signal transduction is a complex process that transmits extra- PTPs belong to a superfamily of enzymes that hydrolytically cellular signals effectively through a cascade of events involving remove phosphate groups from proteins (2). -

Supplemental Material
Supplemental Table 1. Genes activated by alcohol in cultured cortical neurons, as assessed by micro-array analysis. Gene Description Genbank Acc No Folds of increase Gpnmb glycoprotein (transmembrane) nmb NM_053110 2.58 Lyzs lysozyme NM_017372 2.36 Gpnmb glycoprotein (transmembrane) nmb NM_053110 2.33 Gpnmb glycoprotein (transmembrane) nmb NM_053110 2.27 Gpm6a glycoprotein m6a NM_153581 2.05 Mtap1b microtubule-associated protein 1 B NM_008634 2.00 Gfap glial fibrillary acidic protein NM_010277 1.94 C1qg complement component 1, q subcomponent, C chain NM_007574 1.90 C1qb complement component 1, q subcomponent, beta polypeptide, mRNA NM_009777 1.87 Laptm5 lysosomal-associated protein transmembrane 5 NM_010686 1.82 Apoc1 apolipoprotein C-I NM_007469 1.81 Lgals3 lectin, galactose binding, soluble 3 NM_010705 1.81 Fcer1g Fc receptor, IgE, high affinity I, gamma polypeptide NM_010185 1.81 Cd68 CD68 antigen NM_009853 1.81 Apoe apolipoprotein E NM_009696 1.76 C1qa complement component 1, q subcomponent, alpha polypeptide NM_007572 1.75 Lgmn legumain NM_011175 1.74 Msr2 macrophage scavenger receptor 2 NM_030707 1.72 Trem2 triggering receptor expressed on myeloid cells 2 NM_031254 1.72 Serpina3n serine (or cysteine) peptidase inhibitor, clade A, member 3N NM_009252 1.71 Igf1 insulin-like growth factor 1, transcript variant 1 NM_010512 1.71 Ctsz cathepsin Z NM_022325 1.71 Adfp adipose differentiation related protein NM_007408 1.69 Pdgfra platelet derived growth factor receptor, alpha polypeptide NM_011058 1.67 Mmp12 matrix metallopeptidase 12 NM_008605 -
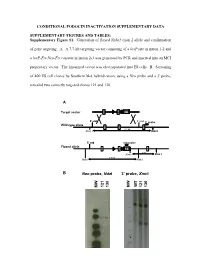
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex. -

Protein Tyrosine Phosphatases in Health and Disease Wiljan J
REVIEW ARTICLE Protein tyrosine phosphatases in health and disease Wiljan J. A. J. Hendriks1, Ari Elson2, Sheila Harroch3, Rafael Pulido4, Andrew Stoker5 and Jeroen den Hertog6,7 1 Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands 2 Department of Molecular Genetics, The Weizmann Institute of Science, Rehovot, Israel 3 Department of Neuroscience, Institut Pasteur, Paris, France 4 Centro de Investigacio´ nPrı´ncipe Felipe, Valencia, Spain 5 Neural Development Unit, Institute of Child Health, University College London, UK 6 Hubrecht Institute, KNAW & University Medical Center Utrecht, The Netherlands 7 Institute of Biology Leiden, Leiden University, The Netherlands Keywords Protein tyrosine phosphatases (PTPs) represent a super-family of enzymes bone morphogenesis; hereditary disease; that play essential roles in normal development and physiology. In this neuronal development; post-translational review, we will discuss the PTPs that have a causative role in hereditary modification; protein tyrosine phosphatase; diseases in humans. In addition, recent progress in the development and synaptogenesis analysis of animal models expressing mutant PTPs will be presented. The Correspondence impact of PTP signaling on health and disease will be exemplified for the J. den Hertog, Hubrecht Institute, fields of bone development, synaptogenesis and central nervous system dis- Uppsalalaan 8, 3584 CT Utrecht, eases. Collectively, research on PTPs since the late 1980’s yielded the The Netherlands cogent view that development of PTP-directed -

Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction Analysis of Melanoma Progression-Associated Genes
ANTICANCER RESEARCH 27: 1301-1308 (2007) Real-time Quantitative Reverse Transcriptase-polymerase Chain Reaction Analysis of Melanoma Progression-associated Genes MAIRIN RAFFERTY1, WILLIAM J. FALLER1, CATHERINE MOSS3, JANET MCCORMACK3, GUNHILD M. MÃLANDSMO4, DAVID J. EASTY2 and WILLIAM M. GALLAGHER1 1UCD School of Biomolecular and Biomedical Science, 2UCD School of Medicine and Medical Science and 3Transcriptomics Core, UCD Conway Institute, University College Dublin, Ireland; 4Department of Tumor Biology, Institute for Cancer Research, The Norwegian Radium Hospital, Montebello, 0310 Oslo, Norway Abstract. Background: Melanoma is an aggressive disease signalling pathways in melanoma (4-7). These studies have that spreads quickly and is resistant to most therapeutic agents. identified cohorts of genes that facilitate the differentiation In an effort to provide insight into the molecular basis of of benign nevi from malignant melanomas (6), the melanoma progression, the expression of 94 genes in 20 subclassification of metastatic melanomas into distinct metastatic melanomas using a high-throughput real-time subgroups (4, 5) and the prediction of distant metastasis- quantitative RT-PCR assay was analysed. Materials and free survival (7). Moreover, genes associated with Methods: A TaqMan low density array (LDA) was designed progression have also been identified using DNA containing probes/primers directed towards a cohort of genes microarray-based gene expression profile analysis of previously found to be differentially expressed in an isogenic melanoma cell lines (8, 9). cell line model of melanoma progression. For each sample, One of the main stumbling blocks inhibiting melanoma cDNA was prepared and added to the quantitative assay. The studies, however, is the lack of fresh tissue.