Feature Selection and Classification of MAQC-II Breast Cancer and Multiple Myeloma Microarray Gene Expression Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Data
Figure 2S 4 7 A - C 080125 CSCs 080418 CSCs - + IFN-a 48 h + IFN-a 48 h + IFN-a 72 h 6 + IFN-a 72 h 3 5 MRFI 4 2 3 2 1 1 0 0 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 7 B 13 080125 FBS - D 080418 FBS - + IFN-a 48 h 12 + IFN-a 48 h + IFN-a 72 h + IFN-a 72 h 6 080125 FBS 11 10 5 9 8 4 7 6 3 MRFI 5 4 2 3 2 1 1 0 0 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 Molecule Molecule FIGURE 4S FIGURE 5S Panel A Panel B FIGURE 6S A B C D Supplemental Results Table 1S. Modulation by IFN-α of APM in GBM CSC and FBS tumor cell lines. Molecule * Cell line IFN-α‡ HLA β2-m# HLA LMP TAP1 TAP2 class II A A HC§ 2 7 10 080125 CSCs - 1∞ (1) 3 (65) 2 (91) 1 (2) 6 (47) 2 (61) 1 (3) 1 (2) 1 (3) + 2 (81) 11 (80) 13 (99) 1 (3) 8 (88) 4 (91) 1 (2) 1 (3) 2 (68) 080125 FBS - 2 (81) 4 (63) 4 (83) 1 (3) 6 (80) 3 (67) 2 (86) 1 (3) 2 (75) + 2 (99) 14 (90) 7 (97) 5 (75) 7 (100) 6 (98) 2 (90) 1 (4) 3 (87) 080418 CSCs - 2 (51) 1 (1) 1 (3) 2 (47) 2 (83) 2 (54) 1 (4) 1 (2) 1 (3) + 2 (81) 3 (76) 5 (75) 2 (50) 2 (83) 3 (71) 1 (3) 2 (87) 1 (2) 080418 FBS - 1 (3) 3 (70) 2 (88) 1 (4) 3 (87) 2 (76) 1 (3) 1 (3) 1 (2) + 2 (78) 7 (98) 5 (99) 2 (94) 5 (100) 3 (100) 1 (4) 2 (100) 1 (2) 070104 CSCs - 1 (2) 1 (3) 1 (3) 2 (78) 1 (3) 1 (2) 1 (3) 1 (3) 1 (2) + 2 (98) 8 (100) 10 (88) 4 (89) 3 (98) 3 (94) 1 (4) 2 (86) 2 (79) * expression of APM molecules was evaluated by intracellular staining and cytofluorimetric analysis; ‡ cells were treatead or not (+/-) for 72 h with 1000 IU/ml of IFN-α; # β-2 microglobulin; § β-2 microglobulin-free HLA-A heavy chain; ∞ values are indicated as ratio between the mean of fluorescence intensity of cells stained with the selected mAb and that of the negative control; bold values indicate significant MRFI (≥ 2). -

(12) United States Patent (10) Patent No.: US 9.284.609 B2 Tomlins Et Al
USOO9284609B2 (12) United States Patent (10) Patent No.: US 9.284.609 B2 Tomlins et al. (45) Date of Patent: Mar. 15, 2016 (54) RECURRENT GENE FUSIONS IN PROSTATE 4,683, 195 A 7, 1987 Mullis et al. CANCER 4,683.202 A 7, 1987 Mullis et al. 4,800,159 A 1/1989 Mullis et al. 4,873,191 A 10/1989 Wagner et al. (75) Inventors: Scott Tomlins, Ann Arbor, MI (US); 4,965,188 A 10/1990 Mullis et al. Daniel Rhodes, Ann Arbor, MI (US); 4,968,103 A 1 1/1990 McNab et al. Arul Chinnaiyan, Ann Arbor, MI (US); 5,130,238 A 7, 1992 Malek et al. Rohit Mehra, Ann Arbor, MI (US); 5,225,326 A 7/1993 Bresser 5,270,184 A 12/1993 Walker et al. Mark Rubin New York, NY (US); 5,283,174. A 2/1994 Arnold, Jr. et al. Xiao-Wei Sun, New York, NY (US); 5,283,317. A 2/1994 Saifer et al. Sven Perner, Ellwaugen (DE); Charles 5,399,491 A 3, 1995 Kacian et al. Lee, Marlborough, MA (US); Francesca 5,455,166 A 10/1995 Walker Demichelis, New York, NY (US) 5,480,784. A 1/1996 Kacian et al. s s 5,545,524 A 8, 1996 Trent 5,614,396 A 3/1997 Bradley et al. (73) Assignees: THE BRIGHAMAND WOMENS 5,631, 169 A 5/1997 Lakowicz et al. HOSPITAL, INC., Boston, MA (US); 5,710,029 A 1/1998 Ryder et al. THE REGENTS OF THE 5,776,782 A 7/1998 Tsuji UNIVERSITY OF MICHIGAN, Ann 5,814,447 A 9/1998 Ishiguro et al. -

Quantigene Flowrna Probe Sets Currently Available
QuantiGene FlowRNA Probe Sets Currently Available Accession No. Species Symbol Gene Name Catalog No. NM_003452 Human ZNF189 zinc finger protein 189 VA1-10009 NM_000057 Human BLM Bloom syndrome VA1-10010 NM_005269 Human GLI glioma-associated oncogene homolog (zinc finger protein) VA1-10011 NM_002614 Human PDZK1 PDZ domain containing 1 VA1-10015 NM_003225 Human TFF1 Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) VA1-10016 NM_002276 Human KRT19 keratin 19 VA1-10022 NM_002659 Human PLAUR plasminogen activator, urokinase receptor VA1-10025 NM_017669 Human ERCC6L excision repair cross-complementing rodent repair deficiency, complementation group 6-like VA1-10029 NM_017699 Human SIDT1 SID1 transmembrane family, member 1 VA1-10032 NM_000077 Human CDKN2A cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) VA1-10040 NM_003150 Human STAT3 signal transducer and activator of transcripton 3 (acute-phase response factor) VA1-10046 NM_004707 Human ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) VA1-10047 NM_000737 Human CGB chorionic gonadotropin, beta polypeptide VA1-10048 NM_001017420 Human ESCO2 establishment of cohesion 1 homolog 2 (S. cerevisiae) VA1-10050 NM_197978 Human HEMGN hemogen VA1-10051 NM_001738 Human CA1 Carbonic anhydrase I VA1-10052 NM_000184 Human HBG2 Hemoglobin, gamma G VA1-10053 NM_005330 Human HBE1 Hemoglobin, epsilon 1 VA1-10054 NR_003367 Human PVT1 Pvt1 oncogene homolog (mouse) VA1-10061 NM_000454 Human SOD1 Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

The ETS Family Transcription Factors Etv5 and PU.1 Function in Parallel to Promote Th9 Cell Development
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author J Immunol Manuscript Author . Author manuscript; Manuscript Author available in PMC 2017 September 15. Published in final edited form as: J Immunol. 2016 September 15; 197(6): 2465–2472. doi:10.4049/jimmunol.1502383. The ETS family transcription factors Etv5 and PU.1 function in parallel to promote Th9 cell development Byunghee Koh1,2,*, Matthew M Hufford1,*, Duy Pham1,2,*, Matthew R. Olson1, Tong Wu3, Rukhsana Jabeen1, Xin Sun4, and Mark H. Kaplan1,2 1Department of Pediatrics, Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202 2Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN 46202 3Department of Cellular & Integrative Physiology, Indiana University School of Medicine, Indianapolis, IN 46202 4Laboratory of Genetics, University of Wisconsin-Madison, WI 53706 Abstract The IL-9-secreting Th9 subset of CD4 T helper cells develop in response to an environment containing IL-4 and TGFβ, promoting allergic disease, autoimmunity, and resistance to pathogens. We previously identified a requirement for the ETS family transcription factor PU.1 in Th9 development. In this report we demonstrate that the ETS transcription factor ETV5 promotes IL-9 production in Th9 cells by binding and recruiting histone acetyltransferases to the Il9 locus at sites distinct from PU.1. In cells that are deficient in both PU.1 and ETV5 there is lower IL-9 production than in cells lacking either factor alone. In vivo loss of PU.1 and ETV5 in T cells results in distinct affects on allergic inflammation in the lung, suggesting that these factors function in parallel. -

Supporting Information Targeting Antioxidant Pathways with Ferrocenylated N-Heterocyclic Carbene Supported Gold(I)
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information Targeting Antioxidant Pathways with Ferrocenylated N-Heterocyclic Carbene Supported Gold(I) Complexes in A549 Lung Cancer Cells J. F. Arambula,a* R. McCall,a K. J. Sidoran,b D. Magda,c N. A. Mitchell,d C. W. e,f g g b Bielawski, V. M. Lynch, J. L. Sessler , and K. Arumugam * a Department of Chemistry, Georgia Southern University, Statesboro, Georgia, 30460, USA. b Department of Chemistry, Wright State University, 3640 Colonel Glenn Hwy, Dayton, Ohio, 45435, USA. c Lumiphore, Inc., Berkeley, California, 94710, USA. dDepartment of Health Sciences, Gettysburg College, Gettysburg, PA 17325-1400 e Center for Multidimensional Carbon Materials, Institute for Basic Science, Ulsan 689-798, Republic of Korea. fDepartment of Chemistry and Department of Energy Engineering, Ulsan National Institute of Science and Technology (UNIST), Ulsan 689-798, Republic of Korea g Department of Chemistry, University of Texas at Austin, Austin, Texas, 78712, USA. *E-mail: [email protected]; [email protected] Table of Contents Synthesis S2-S2 X-ray Crystallography S2-S2 Electrochemical Data S3-S4 1H and 13C NMR Spectra S5-S10 UV-Vis Absorption Spectra S11-S11 Biological Assays S12-S13 RNA Microarray Heat Map and Differential Gene Expression S14-S26 References S26-S26 S1 Synthesis An 8 mL screw cap vial equipped with a stir bar was charged with compound 5 (40 mg, 0.039 mmol) and AgBF4. (5.5 mg, 0.084 mmol). Dry CH2Cl2 was added to the vial and the resulting mixture was stirred at 25 °C for 4 h. -
Figure S1. Reverse Transcription‑Quantitative PCR Analysis of ETV5 Mrna Expression Levels in Parental and ETV5 Stable Transfectants
Figure S1. Reverse transcription‑quantitative PCR analysis of ETV5 mRNA expression levels in parental and ETV5 stable transfectants. (A) Hec1a and Hec1a‑ETV5 EC cell lines; (B) Ishikawa and Ishikawa‑ETV5 EC cell lines. **P<0.005, unpaired Student's t‑test. EC, endometrial cancer; ETV5, ETS variant transcription factor 5. Figure S2. Survival analysis of sample clusters 1‑4. Kaplan Meier graphs for (A) recurrence‑free and (B) overall survival. Survival curves were constructed using the Kaplan‑Meier method, and differences between sample cluster curves were analyzed by log‑rank test. Figure S3. ROC analysis of hub genes. For each gene, ROC curve (left) and mRNA expression levels (right) in control (n=35) and tumor (n=545) samples from The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer cohort are shown. mRNA levels are expressed as Log2(x+1), where ‘x’ is the RSEM normalized expression value. ROC, receiver operating characteristic. Table SI. Clinicopathological characteristics of the GSE17025 dataset. Characteristic n % Atrophic endometrium 12 (postmenopausal) (Control group) Tumor stage I 91 100 Histology Endometrioid adenocarcinoma 79 86.81 Papillary serous 12 13.19 Histological grade Grade 1 30 32.97 Grade 2 36 39.56 Grade 3 25 27.47 Myometrial invasiona Superficial (<50%) 67 74.44 Deep (>50%) 23 25.56 aMyometrial invasion information was available for 90 of 91 tumor samples. Table SII. Clinicopathological characteristics of The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer dataset. Characteristic n % Solid tissue normal 16 Tumor samples Stagea I 226 68.278 II 19 5.740 III 70 21.148 IV 16 4.834 Histology Endometrioid 271 81.381 Mixed 10 3.003 Serous 52 15.616 Histological grade Grade 1 78 23.423 Grade 2 91 27.327 Grade 3 164 49.249 Molecular subtypeb POLE 17 7.328 MSI 65 28.017 CN Low 90 38.793 CN High 60 25.862 CN, copy number; MSI, microsatellite instability; POLE, DNA polymerase ε. -

Glycylglycine Plays Critical Roles in the Proliferation of Spermatogonial Stem Cells
3802 MOLECULAR MEDICINE REPORTS 20: 3802-3810, 2019 Glycylglycine plays critical roles in the proliferation of spermatogonial stem cells BO XU1,2*, XIANG WEI1*, MINJIAN CHEN1,2*, KAIPENG XIE3,4, YUQING ZHANG1,2, ZHENYAO HUANG1,2, TIANYU DONG1,2, WEIYUE HU1,2, KUN ZHOU1,2, XIUMEI HAN1,2, XIN WU1 and YANKAI XIA1,2 1State Key Laboratory of Reproductive Medicine, Institute of Toxicology; 2Key Laboratory of Modern Toxicology of Ministry of Education, School of Public Health, Nanjing Medical University, Nanjing, Jiangsu 211166; 3Nanjing Maternity and Child Health Care Institute, 4Department of Women Health Care, Nanjing Maternity and Child Health Care Hospital, Obstetrics and Gynecology Hospital Affiliated to Nanjing Medical University, Nanjing, Jiangsu 210004, P.R. China Received May 9, 2018; Accepted July 9, 2019 DOI: 10.3892/mmr.2019.10609 Abstract. Glial cell line-derived neurotrophic factor (GDNF) properties that distinguish stem cells from somatic cells, is critical for the proliferation of spermatogonial stem cells and SSCs are the only germline stem cells that can undergo (SSCs), but the underlying mechanisms remain poorly under- self-renewal division (1). The balance between proliferation stood. In this study, an unbiased metabolomic analysis was and differentiation is therefore essential to the normal function performed to examine the metabolic modifications in SSCs of SSCs and to maintain male fertility (2). following GDNF deprivation, and 11 metabolites were observed Glial cell line-derived neurotrophic factor (GDNF) is to decrease while three increased. Of the 11 decreased metab- secreted by Sertoli cells, and is an important factor in the olites identified, glycylglycine was observed to significantly cell fate determination of SSCs, which was identified in rescue the proliferation of the impaired SSCs, while no such the year 2000 (3). -

Feature Selection and Classification of MAQC-II Breast Cancer and Multiple Myeloma Microarray Gene Expression Data Qingzhong Liu New Mexico Tech
Florida International University FIU Digital Commons Robert Stempel College of Public Health & Social Department of Epidemiology Work 12-11-2009 Feature Selection and Classification of MAQC-II Breast Cancer and Multiple Myeloma Microarray Gene Expression Data Qingzhong Liu New Mexico Tech Andrew H. Sung New Mexico Tech Zhongxue Chen Department of Epidemiology and Biostatistics, Florida International University Jianzhong Liu QuantumBio, Inc. Xudong Huang Brigham and Women’s Hospital and Harvard Medical School See next page for additional authors Follow this and additional works at: https://digitalcommons.fiu.edu/epidemiology Part of the Medicine and Health Sciences Commons Recommended Citation Liu Q, Sung AH, Chen Z, Liu J, Huang X, et al. (2009) Feature Selection and Classification of MAQC-II Breast Cancer and Multiple Myeloma Microarray Gene Expression Data. PLoS ONE 4(12): e8250. doi:10.1371/journal.pone.0008250 This work is brought to you for free and open access by the Robert Stempel College of Public Health & Social Work at FIU Digital Commons. It has been accepted for inclusion in Department of Epidemiology by an authorized administrator of FIU Digital Commons. For more information, please contact [email protected]. Authors Qingzhong Liu, Andrew H. Sung, Zhongxue Chen, Jianzhong Liu, Xudong Huang, and Youing Deng This article is available at FIU Digital Commons: https://digitalcommons.fiu.edu/epidemiology/12 Feature Selection and Classification of MAQC-II Breast Cancer and Multiple Myeloma Microarray Gene Expression Data Qingzhong Liu1, -

Rnaseq Analysis of Heart Tissue from Mice Treated with Atenolol and Isoproterenol Reveals a Reciprocal Transcriptional Response Andrea Prunotto1,2, Brian J
Prunotto et al. BMC Genomics (2016) 17:717 DOI 10.1186/s12864-016-3059-6 RESEARCH ARTICLE Open Access RNAseq analysis of heart tissue from mice treated with atenolol and isoproterenol reveals a reciprocal transcriptional response Andrea Prunotto1,2, Brian J. Stevenson2, Corinne Berthonneche3, Fanny Schüpfer3, Jacques S. Beckmann1,2,3, Fabienne Maurer3*† and Sven Bergmann1,2,4*† Abstract Background: The transcriptional response to many widely used drugs and its modulation by genetic variability is poorly understood. Here we present an analysis of RNAseq profiles from heart tissue of 18 inbred mouse strains treated with the β-blocker atenolol (ATE) and the β-agonist isoproterenol (ISO). Results: Differential expression analyses revealed a large set of genes responding to ISO (n = 1770 at FDR = 0.0001) and a comparatively small one responding to ATE (n =23atFDR = 0.0001). At a less stringent definition of differential expression, the transcriptional responses to these two antagonistic drugs are reciprocal for many genes, with an overall anti-correlation of r = −0.3. This trend is also observed at the level of most individual strains even though the power to detect differential expression is significantly reduced. The inversely expressed gene sets are enriched with genes annotated for heart-related functions. Modular analysis revealed gene sets that exhibit coherent transcription profiles across some strains and/or treatments. Correlations between these modules and a broad spectrum of cardiovascular traits are stronger than expected by chance. This provides evidence for the overall importance of transcriptional regulation for these organismal responses and explicits links between co-expressed genes and the traits they are associated with. -
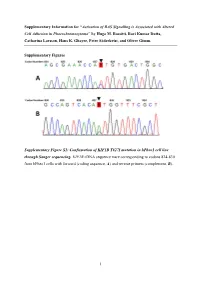
Activation of RAS Signalling Is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M
Supplementary Information for “Activation of RAS Signalling is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M. Rossitti, Ravi Kumar Dutta, Catharina Larsson, Hans K. Ghayee, Peter Söderkvist, and Oliver Gimm. Supplementary Figures Supplementary Figure S1: Confirmation of KIF1B T827I mutation in hPheo1 cell line through Sanger sequencing. KIF1B cDNA sequence trace corresponding to codons 824-830 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 1 Supplementary Figure S2: Confirmation of NRAS Q61K mutation in hPheo1 cell line through Sanger sequencing. NRAS cDNA sequence trace corresponding to codons 56-67 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 2 Supplementary Figure S3: CCND1 gene expression and hPheo1 proliferation. A: Expression of CCND1 mRNA assessed by RT-qPCR and presented as fold change (2-ΔΔCT, mean ± standard error of the mean). B: Cell counts at 1, 2, and 3 days after plating (corresponding to 72, 96 and 120 hours posttransfection, respectively) of control- or siNRAS#1-transfected hPheo1 cells expressed as fold change of the number of cells plated at day 0 (48 hours posttransfection; mean ± standard deviation). All results are from three independent siRNA experiments. 3 Supplementary Tables Supplementary Table S1: List of transcript cluster IDs significantly upregulated in hPheo1 by siNRAS treatment (comparison: siNRAS versus control-transfected hPheo1; ANOVA p < 0.05, FDR < 0.25, fold change < -1.5 or > 1.5). Transcript -

Control of the Physical and Antimicrobial Skin Barrier by an IL-31–IL-1 Signaling Network
The Journal of Immunology Control of the Physical and Antimicrobial Skin Barrier by an IL-31–IL-1 Signaling Network Kai H. Ha¨nel,*,†,1,2 Carolina M. Pfaff,*,†,1 Christian Cornelissen,*,†,3 Philipp M. Amann,*,4 Yvonne Marquardt,* Katharina Czaja,* Arianna Kim,‡ Bernhard Luscher,€ †,5 and Jens M. Baron*,5 Atopic dermatitis, a chronic inflammatory skin disease with increasing prevalence, is closely associated with skin barrier defects. A cy- tokine related to disease severity and inhibition of keratinocyte differentiation is IL-31. To identify its molecular targets, IL-31–dependent gene expression was determined in three-dimensional organotypic skin models. IL-31–regulated genes are involved in the formation of an intact physical skin barrier. Many of these genes were poorly induced during differentiation as a consequence of IL-31 treatment, resulting in increased penetrability to allergens and irritants. Furthermore, studies employing cell-sorted skin equivalents in SCID/NOD mice demonstrated enhanced transepidermal water loss following s.c. administration of IL-31. We identified the IL-1 cytokine network as a downstream effector of IL-31 signaling. Anakinra, an IL-1R antagonist, blocked the IL-31 effects on skin differentiation. In addition to the effects on the physical barrier, IL-31 stimulated the expression of antimicrobial peptides, thereby inhibiting bacterial growth on the three-dimensional organotypic skin models. This was evident already at low doses of IL-31, insufficient to interfere with the physical barrier. Together, these findings demonstrate that IL-31 affects keratinocyte differentiation in multiple ways and that the IL-1 cytokine network is a major downstream effector of IL-31 signaling in deregulating the physical skin barrier.