CLNS1A Antibody (C-Term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap6520b
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

SUPPLEMENTARY TABLES and FIGURE LEGENDS Supplementary
SUPPLEMENTARY TABLES AND FIGURE LEGENDS Supplementary Figure 1. Quantitation of MYC levels in vivo and in vitro. a) MYC levels in cell lines 6814, 6816, 5720, 966, and 6780 (corresponding to first half of Figure 1a in main text). MYC is normalized to tubulin. b) MYC quantitations (normalized to tubulin) for cell lines Daudi, Raji, Jujoye, KRA, KRB, GM, and 6780 corresponding to second half of Figure 1a. c) In vivo MYC quantitations, for mice treated with 0-0.5 ug/ml doxycycline in their drinking water. MYC is normalized to tubulin. d) Quantitation of changing MYC levels during in vitro titration, normalized to tubulin. e) Levels of Odc (normalized to tubulin) follow MYC levels in titration series. Supplementary Figure 2. Evaluation of doxycycline concentration in the plasma of mice treated with doxycycline in their drinking water. Luciferase expressing CHO cells (Tet- off) (Clonethech Inc) that is responsive to doxycycline by turning off luciferase expression was treated with different concentrations of doxycycline in culture. A standard curve (blue line) correlating luciferase activity (y-axis) with treatment of doxycycline (x- axis) was generated for the CHO cell in culture. Plasma from mice treated with different concentrations of doxycycline in their drinking water was separated and added to the media of the CHO cells. Luciferase activity was measured and plotted on the standard curve (see legend box). The actual concentration of doxycycline in the plasma was extrapolated for the luciferase activity measured. The doxycycline concentration 0.2 ng/ml measured in the plasma of mice correlates with 0.05 μg/ml doxycycline treatment in the drinking water of mice, the in vivo threshold for tumor regression. -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Characterization and Proteomic-Transcriptomic Investigation of Monocarboxylate Transporter 6 Knockout Mice
Supplemental material to this article can be found at: http://molpharm.aspetjournals.org/content/suppl/2019/07/10/mol.119.116731.DC1 1521-0111/96/3/364–376$35.00 https://doi.org/10.1124/mol.119.116731 MOLECULAR PHARMACOLOGY Mol Pharmacol 96:364–376, September 2019 Copyright ª 2019 by The American Society for Pharmacology and Experimental Therapeutics Characterization and Proteomic-Transcriptomic Investigation of Monocarboxylate Transporter 6 Knockout Mice: Evidence of a Potential Role in Glucose and Lipid Metabolism s Robert S. Jones, Chengjian Tu, Ming Zhang, Jun Qu, and Marilyn E. Morris Department of Pharmaceutical Sciences, School of Pharmacy and Pharmaceutical Sciences, University at Buffalo, State University of New York, Buffalo, New York (R.S.J., C.T., J.Q., M.E.M.); and New York State Center of Excellence in Bioinformatics and Life Sciences, Buffalo, New York (C.T., M.Z., J.Q.) Received March 21, 2019; accepted June 27, 2019 Downloaded from ABSTRACT Monocarboxylate transporter 6 [(MCT6), SLC16A5] is an or- Mct62/2 mice demonstrated significant changes in 199 genes phan transporter with no known endogenous substrates or in the liver compared with Mct61/1 mice. In silico biological physiological role. Previous in vitro and in vivo experiments pathway analyses revealed significant changes in proteins and molpharm.aspetjournals.org investigated MCT6 substrate/inhibitor specificity in Xenopus genes involved in glucose and lipid metabolism–associated laevis oocytes; however, these data remain limited. Transcrip- pathways. This study is the first to provide evidence for an tomic changes in the livers of mice undergoing different dieting association of Mct6 in the regulation of glucose and lipid schemes have suggested that Mct6 plays a role in glucose metabolism. -

Genome-Wide Transcriptional Analysis of Carboplatin Response in Chemosensitive and Chemoresistant Ovarian Cancer Cells
1605 Genome-wide transcriptional analysis of carboplatin response in chemosensitive and chemoresistant ovarian cancer cells David Peters,1,2 John Freund,2 and Robert L. Ochs2 variety of disease processes, including cancer. Patterns of global gene expression can reveal the molecular pathways 1 Department of Pharmacology and Therapeutics, University of relevant to the disease process and identify potential new Liverpool, United Kingdom and 2Precision Therapeutics, Inc., Pittsburgh, Pennsylvania therapeutic targets. The use of this technology for the molecular classification of cancer was recently shown with the identification of an expression profile that was Abstract predictive of patient outcome for B-cell lymphoma (1). We have recently described an ex vivo chemoresponse In addition, this study showed that histologically similar assay for determining chemosensitivity in primary cultures tumors can be differentiated based on their gene expression of human tumors. In this study, we have extended these profiles. Ultimately, these unique patterns of gene expres- experiments in an effort to correlate chemoresponse data sion may be used as guidelines to direct different modes of with gene expression patterns at the level of transcription. therapy. Primary cultures of cells derived from ovarian carcinomas Although it is widely recognized that patients with the of individual patients (n = 6) were characterized using same histologic stage and grade of cancer respond to the ChemoFx assay and classified as either carboplatin therapies differently, few clinical tests can predict indi- sensitive (n = 3) or resistant (n = 3). Three representa- vidual patient responses. The next great challenge will be tive cultures of cells from each individual tumor were then to use the power of post-genomic technology, including subjected to Affymetrix gene chip analysis (n = 18) using microarray analyses, to correlate gene expression patterns U95A human gene chip arrays. -

Identification of Biomarkers, Pathways and Potential Therapeutic Targets for Heart Failure Using Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.05.455244; this version posted August 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of biomarkers, pathways and potential therapeutic targets for heart failure using bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.08.05.455244; this version posted August 6, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Heart failure (HF) is a complex cardiovascular diseases associated with high mortality. To discover key molecular changes in HF, we analyzed next-generation sequencing (NGS) data of HF. In this investigation, differentially expressed genes (DEGs) were analyzed using limma in R package from GSE161472 of the Gene Expression Omnibus (GEO). Then, gene enrichment analysis, protein-protein interaction (PPI) network, miRNA-hub gene regulatory network and TF-hub gene regulatory network construction, and topological analysis were performed on the DEGs by the Gene Ontology (GO), REACTOME pathway, STRING, HiPPIE, miRNet, NetworkAnalyst and Cytoscape. Finally, we performed receiver operating characteristic curve (ROC) analysis of hub genes. A total of 930 DEGs 9464 up regulated genes and 466 down regulated genes) were identified in HF. -

CLNS1A Rabbit Polyclonal Antibody – TA590737 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA590737 CLNS1A Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: ELISA, IF, IHC, WB Recommended Dilution: WB 1:5000~20000,ELISA 1:100-1:2000 Reactivity: Human Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: DNA immunization. This antibody is specific for the N Terminus Region of the target protein. Formulation: 20 mM Potassium Phosphate, 150 mM Sodium Chloride, pH 7.0 Concentration: 1.01mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Gene Name: chloride nucleotide-sensitive channel 1A Database Link: NP_001284 Entrez Gene 1207 Human P54105 Background: The interaction with Sm proteins inhibits CLNS1A assembly on U RNA and interferes with snRNP biogenesis. CLNS1A inhibits the binding of survival motor neuron protein (SMN) to Sm proteins. May participate in cellular volume control by activation of a swelling-induced chloride conductance pathway. Synonyms: CLCI; CLNS1B; ICln Note: This antibody was generated by SDIX's Genomic Antibody Technology ® (GAT). Learn about GAT Protein Families: Ion Channels: Other This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 3 CLNS1A Rabbit Polyclonal Antibody – TA590737 Product images: HEK293T cells were transfected with the pCMV6- ENTRY control (Cat# [PS100001], Left lane) or pCMV6-ENTRY CLNS1A (Cat# [RC213169], Right lane) cDNA for 48 hrs and lysed. -
Supplementary File 1 (PDF, 306 Kib)
MiR-92a targets Gene Evidence category miRTarBase ID Gene Evidence category miRTarBase ID ITGA5 Strong MIRT003031 CAMKV Weak MIRT048955 ARID4B Strong MIRT004089 QTRT1 Weak MIRT048956 HIPK3 Strong MIRT004090 ASAH1 Weak MIRT048957 MYLIP Strong MIRT004091 SAP30BP Weak MIRT048958 TP63 Strong MIRT004281 RORA Weak MIRT048959 KAT2B Strong MIRT004331 ELAC2 Weak MIRT048960 ESR2 Strong MIRT004563 HNRNPLL Weak MIRT048961 TGFBR2 Strong MIRT004737 ETV6 Weak MIRT048962 BMPR2 Strong MIRT004932 PRRC2B Weak MIRT048963 CPEB2 Strong MIRT005590 LY6G5B Weak MIRT048964 CDH1 Strong MIRT006029 ALKBH4 Weak MIRT048965 BCL2L11 Strong MIRT006148 DAZAP1 Weak MIRT048966 KLF2 Strong MIRT006372 IPO7 Weak MIRT048967 OSBPL2 Strong MIRT049254 CTC1 Weak MIRT048968 MAPRE1 Strong MIRT049331 CHCHD10 Weak MIRT048969 STAT3 Strong MIRT049359 BSG Weak MIRT048970 HIPK1 Strong MIRT049450 RABGAP1 Weak MIRT048971 RFFL Strong MIRT049627 NMD3 Weak MIRT048972 OSBPL8 Strong MIRT049706 MRPL32 Weak MIRT048973 PCGF5 Strong MIRT049737 RPL11 Weak MIRT048974 PHLPP1 Strong MIRT052970 WEE1 Weak MIRT048975 NR1H4 Strong MIRT054133 BAK1 Weak MIRT048976 PTEN Strong MIRT054327 MYH2 Weak MIRT048977 SOCS5 Strong MIRT054643 INSIG1 Weak MIRT048978 RGS5 Strong MIRT437943 GARNL3 Weak MIRT048979 MAPK8 Strong MIRT437964 HMGCR Weak MIRT048980 MAP2K4 Strong MIRT437965 TUBB2B Weak MIRT048981 DUSP10 Strong MIRT438170 RPL24 Weak MIRT048982 CCL8 Strong MIRT438289 HELQ Weak MIRT048983 DNMT1 Strong MIRT438805 MAPK1IP1L Weak MIRT048984 UVRAG Weak MIRT001199 PLA2G4F Weak MIRT048985 THBS1 Weak MIRT005628 HERC1 Weak -

There Are 1393 Genes Significant by SAM with 90Th Percentile Confidence, the False Discovery Rate Among the 1393 Significant Genes Is 0.09968
Class 1 (NPM1 wild-type); Class 2 (NPM1 mutation). There are 1393 genes significant by SAM With 90th percentile confidence, the false discovery rate among the 1393 significant genes is 0.09968 . The delta value used to identify the significant genes is 0.78224 . The fudge factor for standard deviation is computed as 0.06115 . if ratio is >1 gene expression lower in NPM1mut 307 overlapping genes (overlap of genes lower expressed in NPM1mut and putative target genes) if ratio is <1 gene expression higher in NPM1mut rank, based Geom mean of Geom mean of Ratio of on p- ratios in class ratios in class geom Map value 1=wt 2=mut means Clone gene symbol_1 gene symbol_2 Description UG cluster Location 1 1,750 0,395 4,427 IMAGE:382787 TRH TRH TRH || Thyrotropin-releasing hormone || Hs.182231 || AA069596 || || 7200 || 98655 Hs.182231 3 MLLT3 || Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 || 2 1,689 0,551 3,068 IMAGE:783998 MLLT3 MLLT3 Hs.493585 || AA443284 || || 4300 || 116155 Hs.591085 9 P2RY5 || Purinergic receptor P2Y, G-protein coupled, 5 || Hs.123464 || R91539 || purinergic receptor 3 1,866 0,497 3,754 IMAGE:196488 P2RY5 P2RY5 P2Y5=RB intorn-encoded putative G-protei || 10161 || 112029 Hs.123464 13 4 1,678 0,497 3,375 IMAGE:49923 BAALC BAALC BAALC || Brain and acute leukemia, cytoplasmic || Hs.533446 || H28986 || || 79870 || 311379 Hs.533446 8 5 1,611 0,612 2,634 IMAGE:416374 TSPAN13 TM4SF13 TM4SF13 || Tetraspanin 13 || Hs.364544 || W86201 || || 27075 || 226071 Hs.364544 7 6 1,770 0,557 3,179 -

Proteomic Landscape of the Human Choroid–Retinal Pigment Epithelial Complex
Supplementary Online Content Skeie JM, Mahajan VB. Proteomic landscape of the human choroid–retinal pigment epithelial complex. JAMA Ophthalmol. Published online July 24, 2014. doi:10.1001/jamaophthalmol.2014.2065. eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. eFigure 2. Gene ontology (GO) distributions and pathway analysis of human choroid– retinal pigment epithelial (RPE) protein show tissue similarity. eMethods. Tissue collection, mass spectrometry, and analysis. eTable 1. Complete table of proteins identified in the human choroid‐RPE using LC‐ MS/MS. eTable 2. Top 50 signaling pathways in the human choroid‐RPE using MetaCore. eTable 3. Top 50 differentially expressed signaling pathways in the human choroid‐RPE using MetaCore. eTable 4. Differentially expressed proteins in the fovea, macula, and periphery of the human choroid‐RPE. eTable 5. Differentially expressed transcription proteins were identified in foveal, macular, and peripheral choroid‐RPE (p<0.05). eTable 6. Complement proteins identified in the human choroid‐RPE. eTable 7. Proteins associated with age related macular degeneration (AMD). This supplementary material has been provided by the authors to give readers additional information about their work. © 2014 American Medical Association. All rights reserved. 1 Downloaded From: https://jamanetwork.com/ on 09/25/2021 eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. A. The human choroid‐RPE was dissected into fovea, macula, and periphery samples. B. Fractions of proteins were isolated and digested. C. The peptide fragments were analyzed using multi‐dimensional LC‐MS/MS. D. X!Hunter, X!!Tandem, and OMSSA were used for peptide fragment identification. E. Proteins were further analyzed using bioinformatics. -
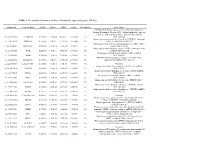
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137.