Integrated Analysis of Cancer Stem Cells-Associated Lncrna-Mirna-Mrna Network for Ovarian Cancer Via Microarray and Gene Expression Omnibus Database
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ovulation-Selective Genes: the Generation and Characterization of an Ovulatory-Selective Cdna Library
531 Ovulation-selective genes: the generation and characterization of an ovulatory-selective cDNA library A Hourvitz1,2*, E Gershon2*, J D Hennebold1, S Elizur2, E Maman2, C Brendle1, E Y Adashi1 and N Dekel2 1Division of Reproductive Sciences, Department of Obstetrics and Gynecology, University of Utah Health Sciences Center, Salt Lake City, Utah 84132, USA 2Department of Biological Regulation, Weizmann Institute of Science, Rehovot, Israel (Requests for offprints should be addressed to N Dekel; Email: [email protected]) *(A Hourvitz and E Gershon contributed equally to this paper) (J D Hennebold is now at Division of Reproductive Sciences, Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, Oregon 97006, USA) Abstract Ovulation-selective/specific genes, that is, genes prefer- (FAE-1) homolog, found to be localized to the inner entially or exclusively expressed during the ovulatory periantral granulosa and to the cumulus granulosa cells of process, have been the subject of growing interest. We antral follicles. The FAE-1 gene is a -ketoacyl-CoA report herein studies on the use of suppression subtractive synthase belonging to the fatty acid elongase (ELO) hybridization (SSH) to construct a ‘forward’ ovulation- family, which catalyzes the initial step of very long-chain selective/specific cDNA library. In toto, 485 clones were fatty acid synthesis. All in all, the present study accom- sequenced and analyzed for homology to known genes plished systematic identification of those hormonally with the basic local alignment tool (BLAST). Of those, regulated genes that are expressed in the ovary in an 252 were determined to be nonredundant. -

Isocitrate Dehydrogenase 1 (NADP+) (I5036)
Isocitrate Dehydrogenase 1 (NADP+), human recombinant, expressed in Escherichia coli Catalog Number I5036 Storage Temperature –20 °C CAS RN 9028-48-2 IDH1 and IDH2 have frequent genetic alterations in EC 1.1.1.42 acute myeloid leukemia4 and better understanding of Systematic name: Isocitrate:NADP+ oxidoreductase these mutations may lead to an improvement of (decarboxylating) individual cancer risk assessment.6 In addition other studies have shown loss of IDH1 in bladder cancer Synonyms: IDH1, cytosolic NADP(+)-dependent patients during tumor development suggesting this may isocitrate dehydrogenase, isocitrate:NADP+ be involved in tumor progression and metastasis.7 oxidoreductase (decarboxylating), Isocitric Dehydrogenase, ICD1, PICD, IDPC, ICDC, This product is lyophilized from a solution containing oxalosuccinate decarboxylase Tris-HCl, pH 8.0, with trehalose, ammonium sulfate, and DTT. Product Description Isocitrate dehydrogenase (NADP+) [EC 1.1.1.42] is a Purity: ³90% (SDS-PAGE) Krebs cycle enzyme, which converts isocitrate to a-ketoglutarate. The flow of isocitrate through the Specific activity: ³80 units/mg protein glyoxylate bypass is regulated by phosphorylation of isocitrate dehydrogenase, which competes for a Unit definition: 1 unit corresponds to the amount of 1 common substrate (isocitrate) with isocitrate lyase. enzyme, which converts 1 mmole of DL-isocitrate to The activity of the enzyme is dependent on the a-ketoglutarate per minute at pH 7.4 and 37 °C (NADP formation of a magnesium or manganese-isocitrate as cofactor). The activity is measured by observing the 2 complex. reduction of NADP to NADPH at 340 nm in the 7 presence of 4 mM DL-isocitrate and 2 mM MnSO4. -

Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’S Biological Network Based on Systematic Pharmacology
ORIGINAL RESEARCH published: 25 June 2021 doi: 10.3389/fphar.2021.601846 Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’s Biological Network Based on Systematic Pharmacology Kailin Yang 1†, Liuting Zeng 1†, Anqi Ge 2†, Yi Chen 1†, Shanshan Wang 1†, Xiaofei Zhu 1,3† and Jinwen Ge 1,4* Edited by: 1 Takashi Sato, Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of 2 Tokyo University of Pharmacy and Life Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, China, Galactophore Department, The First 3 Sciences, Japan Hospital of Hunan University of Chinese Medicine, Changsha, China, School of Graduate, Central South University, Changsha, China, 4Shaoyang University, Shaoyang, China Reviewed by: Hui Zhao, Capital Medical University, China Background: Clinical research found that Hedysarum Multijugum Maxim.-Chuanxiong Maria Luisa Del Moral, fi University of Jaén, Spain Rhizoma Compound (HCC) has de nite curative effect on cerebral ischemic diseases, *Correspondence: such as ischemic stroke and cerebral ischemia-reperfusion injury (CIR). However, its Jinwen Ge mechanism for treating cerebral ischemia is still not fully explained. [email protected] †These authors share first authorship Methods: The traditional Chinese medicine related database were utilized to obtain the components of HCC. The Pharmmapper were used to predict HCC’s potential targets. Specialty section: The CIR genes were obtained from Genecards and OMIM and the protein-protein This article was submitted to interaction (PPI) data of HCC’s targets and IS genes were obtained from String Ethnopharmacology, a section of the journal database. -

Identification and Characterization of Genes That Control Fat
Claire D’Andre et al. Journal of Animal Science and Biotechnology 2013, 4:43 http://www.jasbsci.com/content/4/1/43 JOURNAL OF ANIMAL SCIENCE AND BIOTECHNOLOGY RESEARCH Open Access Identification and characterization of genes that control fat deposition in chickens Hirwa Claire D’Andre1,3*, Wallace Paul2, Xu Shen3, Xinzheng Jia3, Rong Zhang3, Liang Sun3 and Xiquan Zhang3 Abstract Background: Fat deposits in chickens contribute significantly to meat quality attributes such as juiciness, flavor, taste and other organoleptic properties. The quantity of fat deposited increases faster and earlier in the fast- growing chickens than in slow-growing chickens. In this study, Affymetrix Genechip® Chicken Genome Arrays 32773 transcripts were used to compare gene expression profiles in liver and hypothalamus tissues of fast-growing and slow-growing chicken at 8 wk of age. Real-time RT-PCR was used to validate the differential expression of genes selected from the microarray analysis. The mRNA expression of the genes was further examined in fat tissues. The association of single nucleotide polymorphisms of four lipid-related genes with fat traits was examined in a F2 resource population. Results: Four hundred genes in the liver tissues and 220 genes hypothalamus tissues, respectively, were identified to be differentially expressed in fast-growing chickens and slow-growing chickens. Expression levels of genes for lipid metabolism (SULT1B1, ACSBG2, PNPLA3, LPL, AOAH) carbohydrate metabolism (MGAT4B, XYLB, GBE1, PGM1, HKDC1)cholesttrol biosynthesis (FDPS, LSS, HMGCR, NSDHL, DHCR24, IDI1, ME1) HSD17B7 and other reaction or pro- cesses (CYP1A4, CYP1A1, AKR1B1, CYP4V2, DDO) were higher in the fast-growing White Recessive Rock chickens than in the slow-growing Xinghua chickens. -

MMP-25 Metalloprotease Regulates Innate Immune Response Through NF- Κb Signaling Clara Soria-Valles, Ana Gutiérrez-Fernández, Fernando G
MMP-25 Metalloprotease Regulates Innate Immune Response through NF- κB Signaling Clara Soria-Valles, Ana Gutiérrez-Fernández, Fernando G. Osorio, Dido Carrero, Adolfo A. Ferrando, Enrique Colado, This information is current as M. Soledad Fernández-García, Elena Bonzon-Kulichenko, of September 27, 2021. Jesús Vázquez, Antonio Fueyo and Carlos López-Otín J Immunol 2016; 197:296-302; Prepublished online 3 June 2016; doi: 10.4049/jimmunol.1600094 Downloaded from http://www.jimmunol.org/content/197/1/296 Supplementary http://www.jimmunol.org/content/suppl/2016/06/01/jimmunol.160009 Material 4.DCSupplemental http://www.jimmunol.org/ References This article cites 41 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/197/1/296.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 27, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology MMP-25 Metalloprotease Regulates Innate Immune Response through NF-kB Signaling Clara Soria-Valles,* Ana Gutie´rrez-Ferna´ndez,* Fernando G. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

NRF2 and the Ambiguous Consequences of Its Activation During Initiation and the Subsequent Stages of Tumourigenesis
cancers Review NRF2 and the Ambiguous Consequences of Its Activation during Initiation and the Subsequent Stages of Tumourigenesis Holly Robertson 1,2, Albena T. Dinkova-Kostova 1 and John D. Hayes 1,* 1 Jacqui Wood Cancer Centre, Division of Cellular Medicine, Ninewells Hospital and Medical School, University of Dundee, Dundee DD1 9SY, Scotland, UK; [email protected] (H.R.); [email protected] (A.T.D.-K.) 2 Wellcome Trust Sanger Institute, Wellcome Genome Campus, Cambridge CB10 1SA, UK * Correspondence: [email protected]; Tel.: +44-(0)-1382-383182 Received: 30 October 2020; Accepted: 27 November 2020; Published: 2 December 2020 Simple Summary: Transcription factor NRF2 controls expression of antioxidant and detoxification genes. Normally, the activity of NRF2 is tightly controlled in the cell, and is continuously adjusted to ensure that cells are protected against endogenous chemicals and environmental agents that perturb the intracellular antioxidant/pro-oxidant balance (i.e., redox) that must be maintained for them to grow and survive in an appropriate manner. This tight control of NRF2 is achieved by a repressor protein called KEAP1 that perpetually targets NRF2 protein for degradation under normal conditions, but is unable to do so when challenged with oxidants or thiol-reactive chemicals. In the context of cancer, it is well known that drugs that stimulate short-term and reversible activation of NRF2 can provide protection for a limited period against exposure to chemicals that cause cancer. However, it is also becoming widely recognised that permanent hyper-activation of NRF2 resulting from somatic mutations in the gene that encodes NRF2, or in genes associated with its degradation, is frequently observed in certain cancers and associated with poor outcome. -

Development of Potent and Selective Inhibitors of Aldo−Keto Reductase
Article pubs.acs.org/jmc Development of Potent and Selective Inhibitors of Aldo−Keto Reductase 1C3 (Type 5 17β-Hydroxysteroid Dehydrogenase) Based on N-Phenyl-Aminobenzoates and Their Structure−Activity Relationships † § ‡ § † † † ‡ Adegoke O. Adeniji, , Barry M. Twenter, , Michael C. Byrns, Yi Jin, Mo Chen, Jeffrey D. Winkler,*, † and Trevor M. Penning*, † Department of Pharmacology and Center of Excellence in Environmental Toxicology, Perelman School of Medicine, University of Pennsylvania, 130C John Morgan Building, 3620 Hamilton Walk, Philadelphia, Pennsylvania 19104-6084, United States ‡ Department of Chemistry, University of Pennsylvania, 231 South 34th Street, Philadelphia, Pennsylvania 19104, United States *S Supporting Information ABSTRACT: Aldo−keto reductase 1C3 (AKR1C3; type 5 17β-hydroxy- steroid dehydrogenase) is overexpressed in castration resistant prostate cancer (CRPC) and is implicated in the intratumoral biosynthesis of testosterone and 5α-dihydrotestosterone. Selective AKR1C3 inhibitors are required because compounds should not inhibit the highly related AKR1C1 and AKR1C2 isoforms which are involved in the inactivation of 5α-dihydrotestosterone. NSAIDs, N-phenylanthranilates in particular, are potent but nonselective AKR1C3 inhibitors. Using flufenamic acid, 2-{[3-(trifluoromethyl)phenyl]amino}- benzoic acid, as lead compound, five classes of structural analogues were synthesized and evaluated for AKR1C3 inhibitory potency and selectivity. Structure−activity relationship (SAR) studies revealed that a meta-carboxylic acid group relative to the amine conferred pronounced AKR1C3 selectivity without loss of potency, while electron withdrawing groups on the phenylamino B-ring were optimal for AKR1C3 inhibition. Lead compounds did not inhibit COX-1 or COX-2 but blocked the AKR1C3 mediated production of testosterone in LNCaP-AKR1C3 cells. These compounds offer promising leads toward new therapeutics for CRPC. -

Energy Metabolism in IDH1 Wild-Type and IDH1-Mutated Glioblastoma Stem Cells: a Novel Target for Therapy?
cells Review Energy Metabolism in IDH1 Wild-Type and IDH1-Mutated Glioblastoma Stem Cells: A Novel Target for Therapy? Cornelis J.F. van Noorden 1,2,*, Vashendriya V.V. Hira 1, Amber J. van Dijck 2 , Metka Novak 1, Barbara Breznik 1 and Remco J. Molenaar 1,3 1 Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, VeˇcnaPot 111, 1000 Ljubljana, Slovenia; [email protected] (V.V.V.H.); [email protected] (M.N.); [email protected] (B.B.); [email protected] (R.J.M.) 2 Department of Medical Biology, Amsterdam UMC Location Academic Medical Center, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands; [email protected] 3 Department of Medical Oncology, Amsterdam UMC Location Academic Medical Center, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands * Correspondence: [email protected]; Tel.: +31-638-639-561 Abstract: Cancer is a redox disease. Low levels of reactive oxygen species (ROS) are beneficial for cells and have anti-cancer effects. ROS are produced in the mitochondria during ATP production by oxidative phosphorylation (OXPHOS). In the present review, we describe ATP production in primary brain tumors, glioblastoma, in relation to ROS production. Differentiated glioblastoma cells mainly use glycolysis for ATP production (aerobic glycolysis) without ROS production, whereas glioblastoma stem cells (GSCs) in hypoxic periarteriolar niches use OXPHOS for ATP and ROS production, which is modest because of the hypoxia and quiescence of GSCs. In a significant proportion of glioblastoma, Citation: van Noorden, C.J.F.; Hira, isocitrate dehydrogenase 1 (IDH1) is mutated, causing metabolic rewiring, and all cancer cells use V.V.V.; van Dijck, A.J.; Novak, M.; OXPHOS for ATP and ROS production. -

Methylation-Dependent Tissue Factor Suppression Contributes to the Reduced Malignancy of IDH1 Mutant Gliomas
Author Manuscript Published OnlineFirst on September 28, 2018; DOI: 10.1158/1078-0432.CCR-18-1222 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Unruh et al. Methylation-dependent Tissue Factor suppression contributes to the reduced malignancy of IDH1 mutant gliomas 1 1 2 1 1 Dusten Unruh, Snezana Mirkov, Brian Wray, Michael Drumm, Jonathan Lamano, Yuping D Li,1 Qazi F. Haider,1 Rodrigo Javier,1 Kathleen McCortney,1 Amanda Saratsis,1 Denise M. Scholtens,3 Jann N. Sarkaria,4 C. David James,1 Craig Horbinski1,5 1Department of Neurological Surgery, Northwestern University, Chicago, IL, 60611, USA 2Department of Biochemistry and Molecular Genetics, Northwestern University, Chicago, IL, 60611, USA 3Department of Preventive Medicine, Northwestern University, Chicago, IL, 60611, USA 4Department of Radiation Oncology, Mayo Clinic, Rochester, MN, 55905, USA 5Department of Pathology, Northwestern University, Chicago, IL, 60611, USA RUNNING TITLE Tissue Factor in IDH1mut and IDH1wt gliomas KEYWORDS Tissue Factor; glioma; isocitrate dehydrogenase; beta-catenin; protease-activated receptor FINANCIAL SUPPORT This work was supported by NIH grants K08CA155764 and R01NS102669 (C.H.). D.U. was supported by NIH grants T32CA070085 and F32CA216996. C.H., D.M.S., and C.D.J. were also supported by the Northwestern SPORE in Brain Cancer P50CA221747. The Mayo Clinic Brain Tumor Patient-Derived Xenograft National Resource is supported by Mayo Clinic, the Mayo SPORE in Brain Cancer P50CA108961 and NIH grant R24NS92940. Histology and fluorescent 1 Downloaded from clincancerres.aacrjournals.org on September 23, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on September 28, 2018; DOI: 10.1158/1078-0432.CCR-18-1222 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Transcriptome Analysis of Different Multidrug-Resistant Gastric Carcinoma Cells
in vivo 19: 583-590 (2005) Transcriptome Analysis of Different Multidrug-resistant Gastric Carcinoma Cells STEFFEN HEIM1 and HERMANN LAGE2 1Europroteome AG, Berlin-Hennigsdorf; 2Institute of Pathology, Charité Campus Mitte, Schumannstr. 20/21, D-10117 Berlin, Germany Abstract. Multidrug resistance (MDR) of human cancers is advanced cancer. These therapeutic protocols produce an the major cause of failure of chemotherapy. To better overall response rate of about 20% at best using a single- understand the molecular events associated with the agent, and up to 50% using combination chemotherapy. development of different types of MDR, two different multidrug- Thus, a large number of malignancies are incurable. resistant gastric carcinoma cell lines, the MDR1/P-glycoprotein- Generally, gastrointestinal cancers, including gastric expressing cell line EPG85-257RDB and the MDR1/P- carcinoma, are naturally resistant to many anticancer drugs. glycoprotein-negative cell variant EPG85-257RNOV, as well as Additionally, these tumors are able to develop acquired the corresponding drug-sensitive parental cell line EPG85-257P, drug resistance phenotypes, which include the multidrug were used for analyses of the mRNA expression profiles by resistance (MDR) phenomenon. cDNA array hybridization. Of more than 12,000 genes spotted The MDR phenotype is characterized by simultaneous on the arrays, 156 genes were detected as being significantly resistance of tumor cells to various antineoplastic agents regulated in the cell line EPG85-257RDB in comparison to the which are structurally and functionally unrelated. Besides non-resistant cell variant, and 61 genes were found to be the classical MDR phenotype, mediated by the enhanced differentially expressed in the cell line EPG85-257RNOV. -
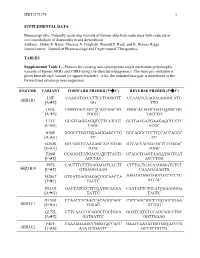
Data Supplement
JPET#173179 1 SUPPLEMENTAL DATA Manuscript title: Naturally occurring variants of human aldo-keto reductases with reduced in vitro metabolism of daunorubicin and doxorubicin Authors: Onkar S. Bains, Thomas A. Grigliatti, Ronald E. Reid, and K. Wayne Riggs Journal name: Journal of Pharmacology and Experimental Therapeutics TABLES Supplemental Table 1—Primers for creating non-synonymous single nucleotide polymorphic variants of human AKRs and CBR4 using site-directed mutagenesis. The base pair mutation is given beneath each variant [in square brackets]. Also, the mutated base pair is underlined in the forward and reverse primer sequences. ENZYME VARIANT FORWARD PRIMER (5'3') REVERSE PRIMER (5'3') I15F CAAGATGCCCTTCCTGGGGTT CCAACCCCAGGAAGGGCATC AKR1B1 [AT] GG TTG H42L CGGGTACCGCCTCATCGACTG GGGCACAGTCGATGAGGCGG [AT] TGCCC TACCCG L73V GCGTGAGGAGGTCTTCATCGT GCTGACGATGAAGACCTCCTC [CG] CAGC ACGC K90E GGGCCTGGTGGAAGGAGCCTG GGCAGGCTCCTTCCACCAGGC [AG] CC CC G204S GCCAGTCCAAAAGCATCGTGG GTCACCACGATGCTTTTGGAC [GA] TGAC TGGC T288I CCAGGATATGACCATCTTACTC GTAGCTGAGTAAGATGGTCAT [CT] AGCTAC ATCCTGG P87S CACTTTCTTTGAGAGATCCCTT CTTTCCTCACAAGGGATCTCT AKR1B10 [CT] GTGAGGAAAG CAAAGAAAGTG M286T GTGATGAGGAGACGGCAACCA GAGTATGGTTGCCGTCTCCTC [TC] TACTC ATCAC N313D GACTATCCCTTCGATGCAGAA CAATATTCTGCATCGAAGGGA [AG] TATTG TAGTC R170H CCAACTTCAACCACAGGCAGC CTCCAGCTGCCTGTGGTTGAA AKR1C1 [GA] TGGAG GTTGG Q172L CTTCAACCGCAGGCTGCTGGA GGATCATCTCCAGCAGCCTGC [AT] GATGATCC GGTTGAAG F46Y CAATAGAAGCCGGGTACCACC GAATCAATATGGTGGTACCCG AKR1C2 [TA] ATATTTGATTC GCTTCTATTG JPET#173179