Transcriptome Analysis of the Curry Tree (Bergera Koenigii L., Rutaceae) During Leaf Development Received: 5 June 2017 Vikram S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Progress and Final Report for Micropropagation of Curry Tree For
Progress and Final Report for Micropropagation of Curry Tree for Nursery Sales and Production of Natural Enemies of the Asian Citrus Psyllid California Citrus Nursery Board - GOD-13 Time Period: January 1, 2013 – December 31, 2013 Project Leader: Kris Godfrey, Contained Research Facility, University of California, 555 Hopkins Road, Davis, CA 95616; Phone: 530-754-2104; Fax: 530-74-8179; Email: [email protected] Cooperating Personnel: David Tricoli, Plant Transformation Facility, 190 Robbins Hall, University of California – Davis, Davis, CA 95616, Phone: 530-752-3766, email: [email protected]; David Morgan, California Dept. of Food and Agriculture, 4500 Glenwood Dr., Bldg. E., Riverside, CA 92501, Phone: 951-328-2642; Email: [email protected] Executive Summary: This project was funded for one year, so research outlined under objective 1 was conducted. The other 2 objectives (consumer acceptance and outreach) were to be performed during year 2 of the project and were not funded. However, we are looking for funding for the outreach component of this project, and the plants generated will be used for Asian citrus psyllid (ACP) rearing by the California Dept. of Food and Agriculture (CDFA). This report summarizes the research accomplished for objective 1, and all studies were conducted by the UC-Davis Plant Transformation Facility. Methods for micropropagation of curry trees were developed using several types of media for both seedlings from seed and nodal explants from mature trees. Establishment of in vitro shoot cultures and multiplication of shoots in vitro has been achieved and improvements in shoot quality were observed by altering the basal salt formulations. -

Show Activity
A Cytochrome-P450-Inhibitor *Unless otherwise noted all references are to Duke, James A. 1992. Handbook of phytochemical constituents of GRAS herbs and other economic plants. Boca Raton, FL. CRC Press. Plant # Chemicals Total PPM Acacia farnesiana Huisache; Cassie; Popinac; Sweet Acacia; Opopanax 2 Achillea millefolium Yarrow; Milfoil 1 Acorus calamus Flagroot; Sweetroot; Sweet Calamus; Myrtle Flag; Calamus; Sweetflag 1 384.0 Agastache rugosa 1 Ageratum conyzoides Mexican ageratum 1 Aloysia citrodora Lemon Verbena 1 Alpinia officinarum Lesser Galangal; Chinese Ginger 1 800.0 Alpinia galanga Siamese Ginger; Languas; Greater Galangal 1 24000.0 Ammi majus Bishop's Weed 2 16000.0 Anacardium occidentale Cashew 1 Anethum graveolens Garden Dill; Dill 1 Angelica dahurica Bai Zhi 2 Angelica archangelica Angelica; Wild Parsnip; Garden Angelica 2 5050.0 Apium graveolens Celery 3 Artemisia dracunculus Tarragon 2 141.0 Boronia megastigma Scented Boronia 1 Calamintha nepeta Turkish Calamint 1 Camellia sinensis Tea 2 Cananga odorata Cananga; Ylang-Ylang 1 Capsicum frutescens Tabasco; Cayenne; Chili; Hot Pepper; Spur Pepper; Red Chili 1 35800.0 Capsicum annuum Cherry Pepper; Cone Pepper; Paprika; Bell Pepper; Sweet Pepper; Green Pepper 2 8000.0 Centaurea calcitrapa Star-Thistle 1 Chenopodium album Lambsquarter 1 Cinnamomum verum Ceylon Cinnamon; Cinnamon 1 20320.0 Cinnamomum camphora Camphor; Ho Leaf 1 Cinnamomum aromaticum Cassia Lignea; Chinese Cassia; Chinesischer Zimtbaum (Ger.); Canela de la China (Sp.); 1 Saigon Cinnamon; Chinazimt (Ger.); Kashia-Keihi -

Show Activity
A Cytotoxic (Colon) *Unless otherwise noted all references are to Duke, James A. 1992. Handbook of phytochemical constituents of GRAS herbs and other economic plants. Boca Raton, FL. CRC Press. Plant # Chemicals Total PPM Aegle marmelos Bael fruit; Bael de India 1 Ammi visnaga Visnaga 1 Ammi majus Bishop's Weed 2 40000.0 Anethum graveolens Garden Dill; Dill 1 Angelica dahurica Bai Zhi 3 Angelica archangelica Angelica; Garden Angelica; Wild Parsnip 3 1902.0 Apium graveolens Celery 3 368.51 Carum carvi Carum; Comino (Sp.); Comino de prado (Sp.); Caraway; Kummel (Ger.) 1 Chenopodium album Lambsquarter 1 Citrus aurantiifolia Lime 1 Coriandrum sativum Cilantro; Chinese Parsley; Coriander 2 Daucus carota Carrot 2 6.0 Dictamnus albus Akgiritotu; Dittany; Gas Plant; Burning Bush; Gazelotu 2 Ficus carica Figueira (Port.); Feigenbaum (Ger.); Figuier Commun (Fr.); Fig; Fico (Ital.); Higo (Sp.); Echte Feige (Ger.); Higuera 2 12100.0 Comun (Sp.) Foeniculum vulgare Fennel 2 2.0 Glehnia littoralis Bei Sha Shen 2 Glycyrrhiza glabra Smooth Licorice; Commom Licorice; Licorice; Licorice-Root 1 Heracleum sphondylium Cow Parsnip; Hogweed; Meadow Parsnip 1 Levisticum officinale Lovage 1 6.0 Limonia acidissima Manzana De Elefante; Elephant Apple; Wood-Apple 1 Murraya koenigii Curry Leaf Tree; Curry Leaf; Indian Curry Tree 1 Pastinaca sativa Parsnip 3 3621.0 Petroselinum crispum Parsley 3 645.5 Peucedanum ostruthium Masterwort 1 Pimpinella anisum Sweet Cumin; Anise 1 Psoralea corylifolia Malaya Tea; Babchi; Black Dot 2 Ruta graveolens Rue 2 Skimmia japonica Japanese Skimmia 1 Zanthoxylum americanum Northern Prickly Ash 3 Zea mays Corn 1 Dr. Duke's Phytochemical and Ethnobotanical Databases Downloaded Tue Sep 28 01:08:32 EDT 2021 National Agricultural Library. -

Commodity Commodity Variety Abelia Abelia Abott Silk
CPC Variety Table COMMODITY COMMODITY VARIETY p. 1/34 [ 7/7/2014 ] COMMODITY COMMODITY VARIETY p. 2/34 [ 7/7/2014 ] ABELIA ABELIA AMARANTHUS AMARANTHUS ABOTT SILK FLOWER AMARYLLIS NAKED LADY/ BELLADONNA ABRONIA SAND VERBENA AMMI GREEN MIST ETC ABUTILON CHINESE LANTERN AMMOBIUM WINGED EVERLASTING ACACIA ACACIA AMPELOPSIS PORCELAIN BERRY ACALYPHA CHENILLE ANAGALLIS PIMPERNEL ACANTHUS BEARS BREECH ANAPHALIS PEARLY EVERLASTING ACER ACER MISC ANCHUSA ANCHUSA ACER BOX ELDER ANEMONE WIND FLOWER ACER JAPANESE MAPLE ANEMOPSIS YERBA MANSA ACEROLA ACEROLA/ BARBADOS CHERRY ANGELICA ANGELICA ACEROLA (MALPIGHIA) P ACEROLA ANGELONIA ANGELONIA ACHIMENES ACHIMENES ANIGOZANTHOS KANGAROO PAW ACONITUM MONKS HOOD ANISACANTHUS HUMMINGBIRD BUSH ACORUS SWEET FLAG ANISE SWEET ANISE ACUBA GOLD DUST PLANT ANISODONTEA CAPE MALLOW ADENOSTEMMA RED SHANKS/CHAMISE ANNONA ANNONA/ CHERIMOYA/ ETC AEOLLANTHUS AEOLLANTHUS ANNONA ATEMOYA AESCHYNANTHUS LIPSTICK PLANT ANNONA CHERIMOYA BOOTH AFRICAN VIOLET SAINTPAULIA AFRICAN VIOLET ANNONA CHERIMOYA DR WHITE AGAPANTHUS LILY OF THE NILE ANNONA P ANNONA/ CHERIMOYA/ ETC AGASTACHE ANISE HYSSOP/HUMMINGBIRD MINT ANTHEMIS ANTHEMIS AGERATUM FLOSS FLOWER ANTHURIUM ANTHURIUM AGONIS AGONIS ANTIRRHINUM SNAPDRAGON AGRETTI AGRETTI APHELANDRA ZEBRA PLANT AGRIMONIA AGRIMONIA APPLE ANNA AGROSTEMMA CORN COCKLE APPLE APPLE MISC AJUGA CARPET BUGLE APPLE ARKANSAS BLACK AKEBIA AKEBIA MISC, CHOCOLATE VINE APPLE BEVERLY HILLS ALBIZIA SILK TREE APPLE CRABAPPLE ALBUCA ALBUCA APPLE DORSETT GOLDEN ALCHEMILLA LADYS MANTLE APPLE EIN SHEMER ALETRIS TRUE UNICORN -

Herbs and Spices Workshop May 2020
Sustainable Living Series - Workshop ‘Herbs and Spices’ They add a zing to your salsa and depth to your curry. Expensive to buy, yet so easy to grow. We grow Herbs and Spices to nurture and stimulate both ourselves and our gardens. The leaves, stems, roots, bark and ‘sap’ of a variety of plants are used to add a bit of vibrance to our creations. The types of herbs you grow will be dependant on your climate and your tastes. On occasion you can grow plants outside their normal climatic region, however be prepared to manipulate its conditions and be more patient, as your harvest times may be longer. You may also find that the flavours are slightly different outside their traditional climatic region. I find... * Sometimes you will eat far more of something than you can grow. * Sometimes it will be far more economic to buy them. * Sometimes you will grow them, just because you can. That’s the fun of gardening! Using Herbs and Spices Think about the types of dishes, preserves, herbal teas, shampoo/conditioners and hand creams you use. What are the main herbs and spices used? Whatever the cuisine you prefer, there will be various ‘staple’ herbs and spices used. A few examples are: Indian: Cumin, Mustard, Turmeric, Ginger Thai: Lemongrass, Kaffir Lime, Chilli, Coriander Moroccan: Paprika, Cardamon, Cinnamon, Preserved Lemon Italian: Garlic, Basil, Sage, Oregano While many are perennials and available year round in our garden, some are annuals and need to be harvested at their peak for later use. Tips on Storing Herbs & Spices •Dry the plant material either by hanging in a dry warm place or using a dehydrator (as per the instructions) •Store in an airtight, clearly labeled container. -

Show Activity
A Antijet-lag *Unless otherwise noted all references are to Duke, James A. 1992. Handbook of phytochemical constituents of GRAS herbs and other economic plants. Boca Raton, FL. CRC Press. Plant # Chemicals Total PPM Ammi majus Bishop's Weed 1 6200.0 Anethum graveolens Dill; Garden Dill 1 Angelica sinensis Dong Quai; Dang Gui; Chinese Angelica; Dang Quai; Dong Gui; Dang Qui 1 Angelica archangelica Angelica; Garden Angelica; Wild Parsnip 1 6810.0 Apium graveolens Celery 1 1043.16 Balanites aegyptiacus dattier sauvage (Fr.); Desert Date; zachun (India); Zachunbaum (Ger.); betu (India); lalo (India); Soapberry 1 Tree; heglik (Fr.); dattier du desert (Fr.); hingotia (India); mirobalano de Egipto (Sp.) Casimiroa edulis White sapote; Matasano 1 Citrus sinensis Orange 1 Citrus limon Lemon 1 Citrus aurantium Bitter Orange; Petitgrain 1 Citrus aurantiifolia Lime 1 Coriandrum sativum Coriander; Cilantro; Chinese Parsley 1 Daucus carota Carrot 1 0.6 Dictamnus albus Akgiritotu; Gazelotu; Dittany; Burning Bush; Gas Plant 1 Ficus carica Fig; Echte Feige (Ger.); Figueira (Port.); Feigenbaum (Ger.); Figuier Commun (Fr.); Fico (Ital.); Higo (Sp.); Higuera 1 14200.0 Comun (Sp.) Foeniculum vulgare Fennel 1 Glehnia littoralis Bei Sha Shen 1 Glycyrrhiza glabra Licorice-Root; Smooth Licorice; Commom Licorice; Licorice 1 Heracleum laciniatum Laciniate Cow-parsnip 1 Levisticum officinale Lovage 1 Limonia acidissima Manzana De Elefante; Wood-Apple; Elephant Apple 1 Murraya koenigii Curry Leaf Tree; Indian Curry Tree; Curry Leaf 1 Pastinaca sativa Parsnip 1 7600.0 Petroselinum crispum Parsley 1 4000.0 Pimpinella anisum Sweet Cumin; Anise 1 Ruta graveolens Rue 1 Skimmia arborescens Skimmia 1 Dr. Duke's Phytochemical and Ethnobotanical Databases Downloaded Fri Oct 01 14:48:13 EDT 2021 National Agricultural Library. -

Newsletter May 1990
90-29 NEWSLETTER MAY 1990 TA,ITPA BAY CHAPTER of The ffirH RARE FRUIT COUNCIL INTERNATIONAL, lnc. EDITORIAL COMMITTEE: BOB HEATH NEIdSLTTTTR I4AIL ADDRESS: ARNOLD &LILLIAN STARK THERESA HEATH 6305 TUREKA SPR I NGS RD. ARNOLD STARK TAMPA FL 33 610 L ILL IAN STARK PRES I DENT: JULES COHAN CHAPTER MAIL ADDRESS: 313 PRUETT RD., SEFFNER FL 33584 ( INCLUDING RENEWALS) MEETINGS ARE HELD THE Znd SUNDAY OF THE MONTH Ar 2:00 P.M. NEXT MEETI NG MAY 13, 1gg0 MEETI NG PLACE. HILLSBOROUGH COUNTY AGRICULTURAL BUSINESS CTNTER (COUNTY AG. AGENTS' BUILDING, SEFFNER) Take I-4 to Exit B South, go past 'l S.R. 579, traffic ight at u.s. gz 'i ntersection. Build'i ng is less than l/Z mile on left (east) sideof u.s.92. usepark'i nglot. Meeting room is in rear of bu'i lding. Main door w'i ll prob ab'ly be 1ock ed. l^Jal k aroun d. PROGRAM " GENE J0YNER'S tal k i s an annu al tradi ti on, wh i ch we i n trociu ce wi th poeti c ren d j ti on . The topic for the day is fruit of Guatenrala, a subject on which he's a well known scholar. The Jore that he speaks i s nct wri tten in tom€S, but placti cal knowl edge of berri es and pomes. His slides are terrific, his talk is first rate. So be sure to join uS, and don't miss th.i s date. EOHGRATULAIIOilS! Congratulations to Louis Zoehrer and Jules Cohan for the article on the Spring Hill club which appeared in the Hernando County section of'the Tampa Tribune. -

Florida Coonties and Atala Butterflies1
Archival copy: for current recommendations see http://edis.ifas.ufl.edu or your local extension office. ENH117 Florida Coonties and Atala Butterflies1 Daniel F. Culbert2 Sunshine State gardeners have rediscovered the Coontie Relatives Florida coontie (Figure 1) as a native plant well adapted to Florida yards. Its increased use in The coontie, an unusual Florida native, is a landscapes has encouraged the presence of the rare cycad—a "living fossil." These primitive plants were a atala butterfly, which uses coontie as a larval host dominant form of plant life during the dinosaur age. plant. Landscapers and homeowners can encourage Other introduced cycads commonly grown in Florida either the plant or the butterfly by following the are: suggestions in this publication. • Cardboard plant/Mexican zamia (Zamia furfuracea) http://hort.ifas.ufl.edu/shrubs/zamfura.pdf • King sago (Cycas revoluta) http://hort.ifas.ufl.edu/shrubs/cycrev.pdf • Queen sago (Cycas rumphii) (Previously called C. circinalis.) http://edis.ifas.ufl.edu/FP161 Coontie Species. There are many different opinions as to the correct name of the species of coontie found growing in Florida. The predominant taxonomic opinion is that there is a single coontie Figure 1. Coonties are a desirable native plant for Florida landscapes. (Photo: Dan Culbert, UF/IFAS) species in Florida (Zamia floridana), while others feel the coontie is represented by several species. Other species names proposed by one or more botanists for the Florida coontie include Z. integrifolia, Z. pumila, and Z. umbrosa. Z. pumila is 1. This document is Fact Sheet ENH 117, a series of the Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. -

Herbal Plant Catalog Fleurizon
Herbal Plants Content • Aerva lanata • Hygrophilia spinosa • Achyranthes aspera • Hemidesmus indicus • Alternanthera sessilis • Ipomea aquatica • Acalypha indica • Justicia adhatoda • Alpinia galanga • Kaempferia galanga • Amaranthus viridis • Lawsonia inermis • Biophytum reinwardtii • Moringa oleifera • Boerhavia diffusa • Muraya koengi • Cardiospermum halicacabum • Osbeckia octandra • Cassia tora • Ocimum basilicum • Centella asiatica • Ocimum sanctum • Costus speciosus • Piper nigrum • Curcuma longa • Pandanas amaryllifolius • Cymbophogon citratus • Talinum paniculatum • Eclipta prostrata • Synsepalum dulcificum “All the plants we produce are used in ‘Ayurvedic’ treatment apart from their edible value. Ayurveda is considered as the oldest healing science that is designed to help people live long, healthy, and well-balanced lives. The basic principle of Ayurveda is to prevent and treat illness by maintaining balance in the body, mind, and consciousness through proper drinking, diet and lifestyle.” Herbal Plants Aerva lanata is a woody, prostrate or succulent, perennial herb in Aerva lanata the Amaranthaceae family of the genus Aerva The whole plant, especially the leaves, is edible. The leaves are commonly used in soup or eaten as a spinach or as a vegetable. Medicinal Uses Leaves -A leaf-decoction is prepared as a gargle for treating sore-throat and used in various complex treatments against guinea-worm. It is used to wash Babies that have become unconscious during an attack of ma- laria or of some other disease .They are washed with a leaf decoction and at the same time smoke from the burning plant is inhaled. The leaf- sap is also used for eye-complaints. An infusion is given to cure diar- rhea and in an unspecified manner at childbirth, and on sores. -

Murraya Koenigii
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN: 2278-3008, p-ISSN:2319-7676. Volume 7, Issue 6 (Sep. – Oct. 2013), PP 15-18 www.iosrjournals.org Murraya koenigii Satish Chand Saini*, Dr. GBS Reddy Shri Venkateshwara University, NH -24, Rajabpur, Gajraula, Distt. Amroha, Pin Code No. 244236 (UP) Ph No +91- 8859500757, 8859500741, Telefax No :+91-0592-229123, www.svu.edu.in, mail: [email protected] Abstract: Medicinal plants or their bioactive compounds have been utilized by developing countries for primary and traditional healthcare system since very long period of time. In several ancient systems of medicine including Ayurveda, Siddha and Unani, Murraya koenigii, a medicinally important herb from mainly Asian origin has vast number of therapeutic applications such as in bronchial disorders, piles, vomiting, skin diseases etc. The medicinal utilities have been described especially for leaf, stem, bark and oil. The well studied pharmacology and phytochemistry of M. koenigii and therapeutic potential of this plant needs to be compiled in form of review. The present review incorporates the description of M. koenigii, its phytochemical constituents and various pharmacological activities of isolated compounds as well as bioactivity of extract studies carried out by various numbers of laboratories. In addition to that, it highlights its potential to be the important nutraceutical for diabetes and cardioprotection. Plants have been used in traditional medicine for several thousand years. India is perhaps the largest producer of medicinal herbs and is rightly called the “Botanical garden of the World”. Murraya koenigii commonly known as Meethi neem, belongs to the family Rutaceae. -
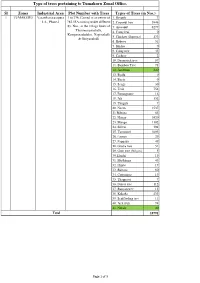
Type of Trees Pertaining to Tumakuru Zonal Office
Type of trees pertaining to Tumakuru Zonal Office. Sl Zones Industrial Area Plot Number with Trees Types of Trees (in Nos.) 1 TUMAKURU Vasanthanarasapura 1 to 236, Carved in an extent of 1. Herculs 7 I.A., Phase-I 782.55A coming under different 2. Coconut tree 3846 Sy. Nos., in the village limits of 3. Arecanut 6277 Thimmarajanahalli, 4. Curry leaf 0 Kempannadodderi, Nagenahalli 5. Chickoo (Sapotaa) 133 & Giriyanahalli 6. Hebevu 51 7. Shishu 0 8. Curry tree 45 9. Cashew 2 10. Drumstick tree 57 11. Bamboo Tree 71 12. Jamblum 340 13. Byala 0 14. Beete 0 15. Seege 30 16. Teak 754 17. Pomogrante 16 18. Jali 132 19. Thugali 7 20. Neem 1537 21 Bilvara 65 22. Honge 1920 23. Mango 1102 24. Sillver 394 25. Tamarind 1003 26. Lemon 29 27. Pappaya 49 28. Guava tree 51 29. Gum tree (Nilgiri) 5 30 Elaichi 19 31. Muthhuga 43 32. Hippe 17 33. Bilvara 80 34. Cottontree 19 35. Thappassi 7 36. Forest tree 112 37. Banyan tree 13 38. Kakada 433 39. Scaffloding tree 11 40. Jack fruit 74 41. Nerale 40 Total 18791 Page 1 of 3 2 Vasanthanarasapura 1 to 657, Carved in an extent of 1. Mango - 5245 I.A., Phase-II 1263.78A coming under different 2. Honge - 1190 Sy. Nos., in the village limits of 3. Bilvara - 5 Yaladdadalu & Nagenahalli 3. Neem - 733 4. Thugali - 174 5. Jali - 169 6. Tamrid - 1064 7. Pomogranate - 0 8. Teak Tree - 1427 9. Seege - 27 10. Jack fruit - 238 11. Beete - 0 12. -

Herb-Spice Chart
Plant List Updated 4/28/2017 Pages !165 © 2017 Rosemary Caspary Key: *cultivated at our garden, ** bought dried, eo- essential oil, ex- extract, io-infused oil Information for educational purposes only and are not intended to replace medical advice, diagnosis, or treatment and does not constitute an endorsement or recommendation by Rosemary Caspary. Consult your health care professional for guidance. Plant Z Contraindicat Companio Ally Enemy Common Scientific o Herb Spice & other Type, Light, Soil, Therapeutic Action/ ion- n Names Name Family n Part of plant: part of plant: Height, Propagation Uses Interactions- Scientific e Caution /Common leaf varies Name Abiu, Pouteria Sapotaceae 9b to Fruit, varieties 1 to 4 Perennial, tree, looks astringent, anti- Caimito caimito Sapodilla 11 seeds, 1 seed similar to canistel, anemic, anti- better, Ztu: dose well in sandy or inflammatory rocky soil; can keep small compact tree, 33’, seed to fruit 3 years Acai Euterpe Perennial, palm tree, oleracea cold and soil sensitive, difficult to grow in S. FL; fruit hard, soften in water *Acoyo, Piper Piperaceae 8a to drink, egg, meat, Perennial, shrub, Leaf: anesthetic, Root Beer auritum Pepper 11 stew, tamales, partial to full shade, stomach disorders Plant, Hoja wrap; Mole Verde moist soil, 6 to 10’, Santa, division, layer Yerba Santa, False Kava Adenophora Adenophor Campanula 7a to cooked Perennial, sun to Root: antifungal, a triphylla ceae, 10b partial shade, moist, cardiotonic, Root, Three- Bellflower leaved 36”, seed expectorant; Ladybells, “Nourish