Evidence Supporting a Viral Origin of the Eukaryotic Nucleus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chapitre Quatre La Spécificité D'hôtes Des Virophages Sputnik
AIX-MARSEILLE UNIVERSITE FACULTE DE MEDECINE DE MARSEILLE ECOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTE THESE DE DOCTORAT Présentée par Morgan GAÏA Né le 24 Octobre 1987 à Aubagne, France Pour obtenir le grade de DOCTEUR de l’UNIVERSITE AIX -MARSEILLE SPECIALITE : Pathologie Humaine, Maladies Infectieuses Les virophages de Mimiviridae The Mimiviridae virophages Présentée et publiquement soutenue devant la FACULTE DE MEDECINE de MARSEILLE le 10 décembre 2013 Membres du jury de la thèse : Pr. Bernard La Scola Directeur de thèse Pr. Jean -Marc Rolain Président du jury Pr. Bruno Pozzetto Rapporteur Dr. Hervé Lecoq Rapporteur Faculté de Médecine, 13385 Marseille Cedex 05, France URMITE, UM63, CNRS 7278, IRD 198, Inserm 1095 Directeur : Pr. Didier RAOULT Avant-propos Le format de présentation de cette thèse correspond à une recommandation de la spécialité Maladies Infectieuses et Microbiologie, à l’intérieur du Master des Sciences de la Vie et de la Santé qui dépend de l’Ecole Doctorale des Sciences de la Vie de Marseille. Le candidat est amené à respecter des règles qui lui sont imposées et qui comportent un format de thèse utilisé dans le Nord de l’Europe permettant un meilleur rangement que les thèses traditionnelles. Par ailleurs, la partie introduction et bibliographie est remplacée par une revue envoyée dans un journal afin de permettre une évaluation extérieure de la qualité de la revue et de permettre à l’étudiant de commencer le plus tôt possible une bibliographie exhaustive sur le domaine de cette thèse. Par ailleurs, la thèse est présentée sur article publié, accepté ou soumis associé d’un bref commentaire donnant le sens général du travail. -

Two Domains of Life, Not Three?
Asgard Archaea Two domains of life, not three? A picture of visual interpretation of the Wagner’s opera Das Rheingold, a part of Richard Wagner’s Der Ring des Nibelungen Loki's Castle is a field of five active hydrothermal vents in the mid-Atlantic Ocean, located at 73 degrees north on the Mid-Atlantic Ridge between Greenland and Norway at a depth of 2,352 metres The vents were discovered in 2008 by a multinational scientific expedition of the university of Bergen, and are the most northerly black smokers to date. The five active chimneys of Loki's Castle are venting water as hot as 300 °C and sit on a vast mound of sulfide minerals. The vent field was given the name Loki's Castle as its shape reminded its discoverers of a fantasy castle. The reference is to the ancient Norse god of trickery, Loki. The top three feet (1 m) of a vent chimney almost 40 feet (12 m) tall at Loki's Castle in mid-July 2008. Visible at left is the arm of a remotely operated vehicle, reaching in to take fluid samples. Loki’s Castle Wents Loki's Castle - a field of five active hydrothermal vents in the mid-Atlantic Ocean - at 73 degrees north on the Mid-Atlantic Ridge - at a depth of 2,352 meters In Norse mythology, Loki is a cunning trickster who has the ability to change his shape and sex. Loki is represented as the companion of the great gods Odin and Thor. Preliminary observations have shown the warm area around the Loki's Castle vents to be alive with diverse and apparently unique microorganisms, unlike vent communities observed elsewhere. -

A Persistent Giant Algal Virus, with a Unique Morphology, Encodes An
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.30.228163; this version posted January 13, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A persistent giant algal virus, with a unique morphology, encodes an 2 unprecedented number of genes involved in energy metabolism 3 4 Romain Blanc-Mathieu1,2, Håkon Dahle3, Antje Hofgaard4, David Brandt5, Hiroki 5 Ban1, Jörn Kalinowski5, Hiroyuki Ogata1 and Ruth-Anne Sandaa6* 6 7 1: Institute for Chemical Research, Kyoto University, Gokasho, Uji, 611-0011, Japan 8 2: Laboratoire de Physiologie Cellulaire & Végétale, CEA, Univ. Grenoble Alpes, 9 CNRS, INRA, IRIG, Grenoble, France 10 3: Department of Biological Sciences and K.G. Jebsen Center for Deep Sea Research, 11 University of Bergen, Bergen, Norway 12 4: Department of Biosciences, University of Oslo, Norway 13 5: Center for Biotechnology, Universität Bielefeld, Bielefeld, 33615, Germany 14 6: Department of Biological Sciences, University of Bergen, Bergen, Norway 15 *Corresponding author: Ruth-Anne Sandaa, +47 55584646, [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.07.30.228163; this version posted January 13, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 16 Abstract 17 Viruses have long been viewed as entities possessing extremely limited metabolic 18 capacities. -
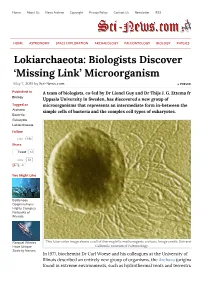
Lokiarchaeota: Biologists Discover 'Missing Link' Microorganism
Home About Us News Archive Copyright Privacy Policy Contact Us Newsletter RSS HOME ASTRONOMY SPACE EXPLORATION ARCHAEOLOGY PALEONTOLOGY BIOLOGY PHYSICS Lokiarchaeota: Biologists Discover ‘Missing Link’ Microorganism May 7, 2015 by Sci-News.com « PREVIOUS Published in A team of biologists, co-led by Dr Lionel Guy and Dr Thijs J. G. Ettema from Biology Uppsala University in Sweden, has discovered a new group of Tagged as microorganisms that represents an intermediate form in-between the Archaea simple cells of bacteria and the complex cell types of eukaryotes. Bacteria Eukaryote Lokiarchaeota Follow Like 16k Share Tweet 12 Like 58 41 You Might Like Bottlenose Dolphins Form Highly Complex Networks of Friends Rorqual Whales This false-color image shows a cell of thermophilic methanogenic archaea. Image credit: University of Have Unique California Museum of Paleontology. Stretchy Nerves In 1977, biochemist Dr Carl Woese and his colleagues at the University of Illinois described an entirely new group of organisms, the Archaea (originally found in extreme environments, such as hydrothermal vents and terrestrial hot springs). The scientists were studying relationships among the prokaryotes using DNA Extinction of sequences, and found that Archaea have distinct molecular characteristics World’s Largest separating them from bacteria as well as from eukaryotes. They proposed that Herbivores May Lead to Empty life can be divided into three domains: Eukaryota, Eubacteria, and Landscapes, Say Archaebacteria. Researchers Despite that archaeal cells were simple and small like bacteria, scientists found that Archaea were more closely related to organisms with complex cell types, a group collectively known as ‘eukaryotes.’ This observation has puzzled Sichuan Bush biologists for years. -

Genomic Exploration of Individual Giant Ocean Viruses
The ISME Journal (2017) 11, 1736–1745 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej ORIGINAL ARTICLE Genomic exploration of individual giant ocean viruses William H Wilson1,2, Ilana C Gilg1, Mohammad Moniruzzaman3, Erin K Field1,4, Sergey Koren5, Gary R LeCleir3, Joaquín Martínez Martínez1, Nicole J Poulton1, Brandon K Swan1,6, Ramunas Stepanauskas1 and Steven W Wilhelm3 1Bigelow Laboratory for Ocean Sciences, Boothbay, ME, USA; 2School of Marine Science and Engineering, Plymouth University, Plymouth, UK; 3Department of Microbiology, The University of Tennessee, Knoxville, TN, USA; 4Department of Biology, Howell Science Complex, East Carolina University, Greenville, NC, USA; 5Genome Informatics Section, Computational and Statistical Genomics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA and 6National Biodefense Analysis and Countermeasures Center, Frederick, MD, USA Viruses are major pathogens in all biological systems. Virus propagation and downstream analysis remains a challenge, particularly in the ocean where the majority of their microbial hosts remain recalcitrant to current culturing techniques. We used a cultivation-independent approach to isolate and sequence individual viruses. The protocol uses high-speed fluorescence-activated virus sorting flow cytometry, multiple displacement amplification (MDA), and downstream genomic sequencing. We focused on ‘giant viruses’ that are readily distinguishable by flow cytometry. From a single- milliliter sample of seawater collected from off the dock at Boothbay Harbor, ME, USA, we sorted almost 700 single virus particles, and subsequently focused on a detailed genome analysis of 12. A wide diversity of viruses was identified that included Iridoviridae, extended Mimiviridae and even a taxonomically novel (unresolved) giant virus. -

Origin and Evolution of Eukaryotic Compartmentalization
TESI DOCTORAL UPF 2016 Origin and evolution of eukaryotic compartmentalization Alexandros Pittis Director Dr. Toni Gabaldon´ Comparative Genomics Group Bioinformatics and Genomics Department Centre for Genomic Regulation (CRG) To my father Stavros for the PhD he never did Acknowledgments Looking back at the years of my PhD in the Comparative Genomics group at CRG, I feel it was equally a process of scientific, as well as personal development. All this time I was very privileged to be surrounded by people that offered me their generous support in both fronts. And they were available in the very moments that I was fighting more myself than the unsolvable (anyway) comparative genomics puzzles. I hope that in the future I will have many times the chance to express them my appreciation, way beyond these few words. First, to Toni, my supervisor, for all his support, and patience, and confidence, and respect, especially at the moments that things did not seem that promising. He offered me freedom, to try, to think, to fail, to learn, to achieve that few enjoy during their PhD, and I am very thankful to him. Then, to my good friends and colleagues, those that I found in the group already, and others that joined after me. A very special thanks to Marinita and Jaime, for all their valuable time, and guidance and paradigm, which nevertheless I never managed to follow. They have both marked my phylogenomics path so far and I cannot escape. To Les and Salvi, my fellow students and pals at the time for all that we shared; to Gab and Fran and Dam, for -

Diversity and Evolution of Viral Pathogen Community in Cave Nectar Bats (Eonycteris Spelaea)
viruses Article Diversity and Evolution of Viral Pathogen Community in Cave Nectar Bats (Eonycteris spelaea) Ian H Mendenhall 1,* , Dolyce Low Hong Wen 1,2, Jayanthi Jayakumar 1, Vithiagaran Gunalan 3, Linfa Wang 1 , Sebastian Mauer-Stroh 3,4 , Yvonne C.F. Su 1 and Gavin J.D. Smith 1,5,6 1 Programme in Emerging Infectious Diseases, Duke-NUS Medical School, Singapore 169857, Singapore; [email protected] (D.L.H.W.); [email protected] (J.J.); [email protected] (L.W.); [email protected] (Y.C.F.S.) [email protected] (G.J.D.S.) 2 NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore 119077, Singapore 3 Bioinformatics Institute, Agency for Science, Technology and Research, Singapore 138671, Singapore; [email protected] (V.G.); [email protected] (S.M.-S.) 4 Department of Biological Sciences, National University of Singapore, Singapore 117558, Singapore 5 SingHealth Duke-NUS Global Health Institute, SingHealth Duke-NUS Academic Medical Centre, Singapore 168753, Singapore 6 Duke Global Health Institute, Duke University, Durham, NC 27710, USA * Correspondence: [email protected] Received: 30 January 2019; Accepted: 7 March 2019; Published: 12 March 2019 Abstract: Bats are unique mammals, exhibit distinctive life history traits and have unique immunological approaches to suppression of viral diseases upon infection. High-throughput next-generation sequencing has been used in characterizing the virome of different bat species. The cave nectar bat, Eonycteris spelaea, has a broad geographical range across Southeast Asia, India and southern China, however, little is known about their involvement in virus transmission. -

A Distinct Lineage of Giant Viruses Brings a Rhodopsin Photosystem to Unicellular Marine Predators
A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators David M. Needhama,1, Susumu Yoshizawab,1, Toshiaki Hosakac,1, Camille Poiriera,d, Chang Jae Choia,d, Elisabeth Hehenbergera,d, Nicholas A. T. Irwine, Susanne Wilkena,2, Cheuk-Man Yunga,d, Charles Bachya,3, Rika Kuriharaf, Yu Nakajimab, Keiichi Kojimaf, Tomomi Kimura-Someyac, Guy Leonardg, Rex R. Malmstromh, Daniel R. Mendei, Daniel K. Olsoni, Yuki Sudof, Sebastian Sudeka, Thomas A. Richardsg, Edward F. DeLongi, Patrick J. Keelinge, Alyson E. Santoroj, Mikako Shirouzuc, Wataru Iwasakib,k,4, and Alexandra Z. Wordena,d,4 aMonterey Bay Aquarium Research Institute, Moss Landing, CA 95039; bAtmosphere & Ocean Research Institute, University of Tokyo, Chiba 277-8564, Japan; cLaboratory for Protein Functional & Structural Biology, RIKEN Center for Biosystems Dynamics Research, Yokohama, Kanagawa 230-0045, Japan; dOcean EcoSystems Biology Unit, GEOMAR Helmholtz Centre for Ocean Research, 24105 Kiel, Germany; eDepartment of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; fGraduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama 700-8530, Japan; gLiving Systems Institute, School of Biosciences, College of Life and Environmental Sciences, University of Exeter, Exeter EX4 4SB, United Kingdom; hDepartment of Energy Joint Genome Institute, Walnut Creek, CA 94598; iDaniel K. Inouye Center for Microbial Oceanography, University of Hawaii, Manoa, Honolulu, HI 96822; jDepartment of Ecology, Evolution and Marine Biology, University of California, Santa Barbara, CA 93106; and kDepartment of Biological Sciences, Graduate School of Science, University of Tokyo, Tokyo 113-0032, Japan Edited by W. Ford Doolittle, Dalhousie University, Halifax, Canada, and approved August 8, 2019 (received for review May 27, 2019) Giant viruses are remarkable for their large genomes, often rivaling genomes that range up to 2.4 Mb (Fig. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

Spatial Separation of Ribosomes and DNA in Asgard Archaeal Cells ✉ ✉ Burak Avcı 1,2 , Jakob Brandt3, Dikla Nachmias4, Natalie Elia4, Mads Albertsen 3, Thijs J
www.nature.com/ismej BRIEF COMMUNICATION OPEN Spatial separation of ribosomes and DNA in Asgard archaeal cells ✉ ✉ Burak Avcı 1,2 , Jakob Brandt3, Dikla Nachmias4, Natalie Elia4, Mads Albertsen 3, Thijs J. G. Ettema 2, Andreas Schramm1,5 and Kasper Urup Kjeldsen1 © The Author(s) 2021 The origin of the eukaryotic cell is a major open question in biology. Asgard archaea are the closest known prokaryotic relatives of eukaryotes, and their genomes encode various eukaryotic signature proteins, indicating some elements of cellular complexity prior to the emergence of the first eukaryotic cell. Yet, microscopic evidence to demonstrate the cellular structure of uncultivated Asgard archaea in the environment is thus far lacking. We used primer-free sequencing to retrieve 715 almost full-length Loki- and Heimdallarchaeota 16S rRNA sequences and designed novel oligonucleotide probes to visualize their cells in marine sediments (Aarhus Bay, Denmark) using catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH). Super-resolution microscopy revealed 1–2 µm large, coccoid cells, sometimes occurring as aggregates. Remarkably, the DNA staining was spatially separated from ribosome-originated FISH signals by 50–280 nm. This suggests that the genomic material is condensed and spatially distinct in a particular location and could indicate compartmentalization or membrane invagination in Asgard archaeal cells. The ISME Journal; https://doi.org/10.1038/s41396-021-01098-3 INTRODUCTION cells in marine sediments [8] yet methodical limitations did not The origin of the eukaryotic cell is a major unresolved puzzle in allow to discern single cells and to draw any in-depth conclusion the history of life. -

Archaea and the Origin of Eukaryotes
REVIEWS Archaea and the origin of eukaryotes Laura Eme, Anja Spang, Jonathan Lombard, Courtney W. Stairs and Thijs J. G. Ettema Abstract | Woese and Fox’s 1977 paper on the discovery of the Archaea triggered a revolution in the field of evolutionary biology by showing that life was divided into not only prokaryotes and eukaryotes. Rather, they revealed that prokaryotes comprise two distinct types of organisms, the Bacteria and the Archaea. In subsequent years, molecular phylogenetic analyses indicated that eukaryotes and the Archaea represent sister groups in the tree of life. During the genomic era, it became evident that eukaryotic cells possess a mixture of archaeal and bacterial features in addition to eukaryotic-specific features. Although it has been generally accepted for some time that mitochondria descend from endosymbiotic alphaproteobacteria, the precise evolutionary relationship between eukaryotes and archaea has continued to be a subject of debate. In this Review, we outline a brief history of the changing shape of the tree of life and examine how the recent discovery of a myriad of diverse archaeal lineages has changed our understanding of the evolutionary relationships between the three domains of life and the origin of eukaryotes. Furthermore, we revisit central questions regarding the process of eukaryogenesis and discuss what can currently be inferred about the evolutionary transition from the first to the last eukaryotic common ancestor. Sister groups Two descendants that split The pioneering work by Carl Woese and colleagues In this Review, we discuss how culture- independent from the same node; the revealed that all cellular life could be divided into three genomics has transformed our understanding of descendants are each other’s major evolutionary lines (also called domains): the archaeal diversity and how this has influenced our closest relative. -

On the Occurrence of Cytochrome P450 in Viruses
On the occurrence of cytochrome P450 in viruses David C. Lamba,b,1, Alec H. Follmerc,1, Jared V. Goldstonea,1, David R. Nelsond, Andrew G. Warrilowb, Claire L. Priceb, Marie Y. Truee, Steven L. Kellyb, Thomas L. Poulosc,e,f, and John J. Stegemana,2 aBiology Department, Woods Hole Oceanographic Institution, Woods Hole, MA 02543; bInstitute of Life Science, Swansea University Medical School, Swansea University, Swansea, SA2 8PP Wales, United Kingdom; cDepartment of Chemistry, University of California, Irvine, CA 92697-3900; dDepartment of Microbiology, Immunology and Biochemistry, University of Tennessee Health Science Center, Memphis, TN 38163; eDepartment of Pharmaceutical Sciences, University of California, Irvine, CA 92697-3900; and fDepartment of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3900 Edited by Michael A. Marletta, University of California, Berkeley, CA, and approved May 8, 2019 (received for review February 7, 2019) Genes encoding cytochrome P450 (CYP; P450) enzymes occur A core set of genes involved in viral replication and lysis is most widely in the Archaea, Bacteria, and Eukarya, where they play often conserved in these viruses (10), yet most genes in the giant important roles in metabolism of endogenous regulatory mole- viruses (70–90%) do not have any obvious homolog in existing cules and exogenous chemicals. We now report that genes for virus databases. multiple and unique P450s occur commonly in giant viruses in the Strikingly, many of the giant viruses possess genes coding for Mimiviridae Pandoraviridae , , and other families in the proposed proteins typically involved in cellular processes, including protein order Megavirales. P450 genes were also identified in a herpesvi- translation, DNA repair, and eukaryotic metabolic pathways Ranid herpesvirus 3 Mycobacterium rus ( ) and a phage ( phage previously thought not to occur in viruses (17).