Accelerating the Future of Liquid Biopsy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

797 Circulating Tumor DNA and Circulating Tumor Cells for Cancer
Medical Policy Circulating Tumor DNA and Circulating Tumor Cells for Cancer Management (Liquid Biopsy) Table of Contents • Policy: Commercial • Coding Information • Information Pertaining to All Policies • Policy: Medicare • Description • References • Authorization Information • Policy History • Endnotes Policy Number: 797 BCBSA Reference Number: 2.04.141 Related Policies Biomarkers for the Diagnosis and Cancer Risk Assessment of Prostate Cancer, #336 Policy1 Commercial Members: Managed Care (HMO and POS), PPO, and Indemnity Plasma-based comprehensive somatic genomic profiling testing (CGP) using Guardant360® for patients with Stage IIIB/IV non-small cell lung cancer (NSCLC) is considered MEDICALLY NECESSARY when the following criteria have been met: Diagnosis: • When tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP or invasive biopsy is medically contraindicated), AND • When prior results for ALL of the following tests are not available: o EGFR single nucleotide variants (SNVs) and insertions and deletions (indels) o ALK and ROS1 rearrangements o PDL1 expression. Progression: • Patients progressing on or after chemotherapy or immunotherapy who have never been tested for EGFR SNVs and indels, and ALK and ROS1 rearrangements, and for whom tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP), OR • For patients progressing on EGFR tyrosine kinase inhibitors (TKIs). If no genetic alteration is detected by Guardant360®, or if circulating tumor DNA (ctDNA) is insufficient/not detected, tissue-based genotyping should be considered. Other plasma-based CGP tests are considered INVESTIGATIONAL. CGP and the use of circulating tumor DNA is considered INVESTIGATIONAL for all other indications. 1 The use of circulating tumor cells is considered INVESTIGATIONAL for all indications. -
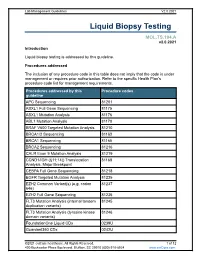
Liquid Biopsy Testing
Lab Management Guidelines V2.0.2021 Liquid Biopsy Testing MOL.TS.194.A v2.0.2021 Introduction Liquid biopsy testing is addressed by this guideline. Procedures addressed The inclusion of any procedure code in this table does not imply that the code is under management or requires prior authorization. Refer to the specific Health Plan's procedure code list for management requirements. Procedures addressed by this Procedure codes guideline APC Sequencing 81201 ASXL1 Full Gene Sequencing 81175 ASXL1 Mutation Analysis 81176 ABL1 Mutation Analysis 81170 BRAF V600 Targeted Mutation Analysis 81210 BRCA1/2 Sequencing 81163 BRCA1 Sequencing 81165 BRCA2 Sequencing 81216 CALR Exon 9 Mutation Analysis 81219 CCND1/IGH (t(11;14)) Translocation 81168 Analysis, Major Breakpoint CEBPA Full Gene Sequencing 81218 EGFR Targeted Mutation Analysis 81235 EZH2 Common Variant(s) (e.g. codon 81237 646) EZH2 Full Gene Sequencing 81236 FLT3 Mutation Analysis (internal tandem 81245 duplication variants) FLT3 Mutation Analysis (tyrosine kinase 81246 domain variants) FoundationOne Liquid CDx 0239U Guardant360 CDx 0242U ©2021 eviCore healthcare. All Rights Reserved. 1 of 12 400 Buckwalter Place Boulevard, Bluffton, SC 29910 (800) 918-8924 www.eviCore.com Lab Management Guidelines V2.0.2021 Procedures addressed by this Procedure codes guideline Hematolymphoid Neoplasm Molecular 81450 Profiling; 5-50 genes IDH1 Mutation Analysis 81120 IDH2 Mutation Analysis 81121 IGH@/BCL2 (t(14;18)) Translocation 81278 Analysis, Major Breakpoint Region (MBR) and Minor Cluster Region (mcr) Breakpoints JAK2 Targeted Mutation Analysis (e.g 81279 exons 12 and 13) JAK2 V617F Targeted Mutation Analysis 81270 KIT Targeted Sequence Analysis 81272 KIT D816 Targeted Mutation Analysis 81273 KRAS Exon 2 Targeted Mutation Analysis 81275 KRAS Targeted Mutation Analysis, 81276 Additional Variants MGMT Promoter Methylation Analysis 81287 MLH1 Sequencing 81292 Molecular Tumor Marker Test 81400 81401 81402 g 81403 n i 81405 t 81406 s e T 81407 81408 y s 81479 p o Molecular Tumor Marker Test 88271 i B MPL Common Variants (e.g. -

Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine Giovanna Rossi1 and Michail Ignatiadis2
Published OnlineFirst May 20, 2019; DOI: 10.1158/0008-5472.CAN-18-3402 Cancer Review Research Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine Giovanna Rossi1 and Michail Ignatiadis2 Abstract New sensitive assays are currently available for the detec- Multiple studies are underway to assess the clinical utility of tion of circulating tumor DNA (ctDNA) and circulating tumor CTC and ctDNA in different settings (treatment-na€ve vs. cells (CTC). However, there remains a need for standardiza- resistant, adjuvant vs. metastatic) and for different treatment tion of preanalytical issues and cross-platform comparison modalities (systemic therapy, surgery, radiation therapy). studies. Liquid biopsies are being evaluated for treatment This review aims to map the challenges that remain to be selection, for monitoring disease response and resistance, for addressed before liquid biopsies can be widely used for cancer tracking minimal residual disease, and for cancer diagnosis. management. Introduction applications of CTCs and ctDNA that are currently being explored are summarized in Fig. 1A. In the era of precision medicine, liquid biopsies are increasingly being studied as a tool that can capture tumor evolution in real time and thus guide systemic treatment. In this article, we will refer to the analysis of circulating tumor DNA (ctDNA) and circulating Challenges Associated with Preanalytical tumor cells (CTC) only and we will not cover other liquid biopsy Issues and the Analytical Validity of Liquid biomarkers such as circulating RNAs, proteins, metabolites, and Biopsy Assays exosomes. For CTC and ctDNA assays, there is a need to standardize Sampling a patient's blood may give information about the preanalytical variables and for cross-platform comparison stud- genomic profile of a given cancer (1–3) and provide an assess- ies. -

Liquid Biopsy Analysis in Clinical Practice: Focus on Lung Cancer
Review Liquid Biopsy Analysis in Clinical Practice: Focus on Lung Cancer Pasquale Pisapia 1 , Francesco Pepe 1, Antonino Iaccarino 1, Roberta Sgariglia 1, Mariantonia Nacchio 1, Gianluca Russo 1 , Gianluca Gragnano 1, Elalah Mosaieby 2, Giancarlo Troncone 1,* and Umberto Malapelle 1 1 Department of Public Health, University of Naples Federico II, 80131 Naples, Italy; [email protected] (P.P.); [email protected] (F.P.); [email protected] (A.I.); [email protected] (R.S.); [email protected] (M.N.); [email protected] (G.R.); [email protected] (G.G.); [email protected] (U.M.) 2 Department of Cellular and Molecular Biology, University of Mazandaran, Mazandaran 48175-866, Iran; [email protected] * Correspondence: [email protected] Abstract: Lung cancer is the leading cause of cancer death worldwide. Despite the emergence of highly effective targeted therapies, up to 30% of advanced stage non-small cell lung cancer (NSCLC) patients do not undergo tissue molecular testing because of scarce tissue availability. Liquid biopsy, on the other hand, offers these patients a valuable opportunity to receive the best treatment options in a timely manner. Indeed, besides being much faster and less invasive than conventional tissue- based analysis, it can also yield specific information about the genetic make-up and evolution of patients’ tumors. However, several issues, including lack of standardized protocols for sample collection, processing, and interpretation, still need to be addressed before liquid biopsy can be Citation: Pisapia, P.; Pepe, F.; fully incorporated into routine oncology practice. Here, we reviewed the most important challenges Iaccarino, A.; Sgariglia, R.; Nacchio, hindering the implementation of liquid biopsy in oncology practice, as well as the great advantages M.; Russo, G.; Gragnano, G.; Mosaieby, E.; Troncone, G.; Malapelle, of this approach for the treatment of NSCLC patients. -

Medtechs Rake in the Venture Cash
January 14, 2021 Medtechs rake in the venture cash Elizabeth Cairns In terms of private investments, 2020 was the year of the liquid biopsy. Looking at the cash raised by private medical device companies last year you would be hard pressed to find any evidence of a pandemic at all. Venture investors poured $6.4bn into the sector, with the particularly strong performance in the second half putting the total markedly above 2019’s. And some of the very biggest investments have already paid off handsomely. The liquid biopsy developers Grail and Thrive hooked nine-figure rounds and were promptly bought for billions of dollars each, by Illumina and Exact Sciences respectively. The VCs backing their fellow cancer blood test specialists Freenome and Caris Life Sciences doubtless also had takeovers in mind. In fact diagnostics companies dominate the top 10 rounds. In addition to the four liquid biopsy groups, Everlywell, Karius and Oxford Nanopore Technologies – two of which are working on tests for Covid-19 – also enjoyed sizeable cash injections. Diagnostics developers, with their relatively cheap and easily scalable technology, are always appealing to VCs, but 2020 was something special. Perhaps the wider environment pushed investors towards these safe bets. But there is a growing consensus that liquid biopsies, though new, are a technology on which oncologists will come to rely. The first US approvals for such tests, from Guardant Health and Roche, were awarded in August, and a third, from Grail, is set for launch without FDA oversight in the second quarter of 2021. Investors’ interest in liquid biopsy plays might also have been influenced by Covid-19. -

Liquid Biopsy: a Family of Possible Diagnostic Tools
diagnostics Review Liquid Biopsy: A Family of Possible Diagnostic Tools Battistelli Michela Department of Biomolecular Sciences, Carlo Bo Urbino University, 61029 Urbino, Italy; [email protected] Abstract: Liquid biopsies could be considered an excellent diagnostic tool, in different physiological or pathological conditions. The possibility of using liquid biopsies for non-invasive clinical purposes is quite an old idea: indeed many years ago it was already being used in the field of non-invasive prenatal tests (NIPT) for autosomal fetal aneuploidy evaluation. In 1997 Lo et al. had identified fetal DNA in maternal plasma and serum, showing that about 10–15% of cfDNA in maternal plasma is derived from the placenta, and biologic fluid represents an important and non-invasive technique to evaluate state diseases and possible therapies. Nowadays, several body fluids, such as blood, urine, saliva and other patient samples, could be used as liquid biopsy for clinical non-invasive evaluation. These fluids contain numerous and various biomarkers and could be used for the evaluation of pathological and non-pathological conditions. In this review we will analyze the different types of liquid biopsy, their potential role in clinical diagnosis and the functional involvement of extracellular vesicles in these fluids as carriers. Keywords: liquid biopsies; extracellular vesicles; tumor dynamic evaluation 1. Introduction Several biological body fluids, such as blood, urine, saliva, breast milk and other Citation: Michela, B. Liquid Biopsy: patient samples, can be used for clinical investigations, because they contain numerous A Family of Possible Diagnostic Tools. biomarkers [1]. The biomarkers found in these liquids can be DNA, RNA, proteins or even Diagnostics 2021, 11, 1391. -

Liquid Biopsies in Solid Cancers: Implementation in a Nordic Healthcare System
cancers Review Liquid Biopsies in Solid Cancers: Implementation in a Nordic Healthcare System Oddmund Nordgård 1,2,*,†, Rakel Brendsdal Forthun 3,4,†, Morten Lapin 1 , Bjørn Henning Grønberg 5,6, Karl Henning Kalland 7,8 , Reidun Kristin Kopperud 7, Liv Cecilie Vestrheim Thomsen 7 , Kjersti Tjensvoll 1, Bjørnar Gilje 1, Bjørn Tore Gjertsen 7,9 and Randi Hovland 3,4 1 Department of Hematology and Oncology, Stavanger University Hospital, 4011 Stavanger, Norway; [email protected] (M.L.); [email protected] (K.T.); [email protected] (B.G.) 2 Department of Chemistry, Bioscience and Environmental Engineering, University of Stavanger, 4021 Stavanger, Norway 3 Department of Medical Genetics, Haukeland University Hospital, 5021 Bergen, Norway; [email protected] (R.B.F.); [email protected] (R.H.) 4 Section of Cancer Genomics, Haukeland University Hospital, 5021 Bergen, Norway 5 Department of Clinical and Molecular Medicine, NTNU, Norwegian University of Science and Technology, 7491 Trondheim, Norway; [email protected] 6 Department of Oncology, St. Olav’s Hospital, Trondheim University Hospital, 7030 Trondheim, Norway 7 Centre for Cancer Biomarkers CCBIO, Department of Clinical Science, University of Bergen, 5021 Bergen, Norway; [email protected] (K.H.K.); [email protected] (R.K.K.); [email protected] (L.C.V.T.); [email protected] (B.T.G.) 8 Department of Microbiology, Haukeland University Hospital, 5021 Bergen, Norway 9 Department of Internal Medicine, Hematology Section, Haukeland University Hospital, 5021 Bergen, Norway Citation: Nordgård, O.; Brendsdal * Correspondence: [email protected] Forthun, R.; Lapin, M.; Grønberg, † Shared first-authorship. -

Guardant Health Q3 2020 Earnings Call
Guardant Health Third Quarter 2020 Earnings Call November 5, 2020 Guardant Health – Third Quarter 2020 Earnings Call, November 5, 2020 C O R P O R A T E P A R T I C I P A N T S Carrie Mendivil, Investor Relations - Gilmartin Group Helmy Eltoukhy, Co-founder and Chief Executive Officer AmirAli Talasaz, Co-founder and President Derek Bertocci, Chief Financial Officer C O N F E R E N C E C A L L P A R T I C I P A N T S Puneet Souda, SVB Tycho Peterson, JPMorgan Doug Schenkel, Cowen Derik de Bruin, Bank of America Brian Weinstein, William Blair Patrick Donnelly, Citigroup Tejas Savant, Morgan Stanley Sung Ji Nam, BTIG P R E S E N T A T I O N Operator Ladies and gentlemen, thank you for standing by, and welcome to the Guardant Health Q3 2020 Earnings Call. At this time all participants are in a listen-only-mode. After the speaker’s presentation, there will be a question-and-answer session. To ask a question during the session, you need to press star, one on your telephone. If you wish to withdraw your question, press the pound key. I would now like to turn the conference over to your speaker today, Carrie Mendivil with Investor Relations. You may begin. Carrie Mendivil 1 ViaVid has made considerable efforts to provide an accurate transcription. There may be material errors, omissions, or inaccuracies in the reporting of the substance of the conference call. This transcript is being made available for information purposes only. -

Microfluidic Chip-Based Cancer Diagnosis and Prediction Of
cancers Review Microfluidic Chip-Based Cancer Diagnosis and Prediction of Relapse by Detecting Circulating Tumor Cells and Circulating Cancer Stem Cells Hyeon-Yeol Cho 1,2,† , Jin-Ha Choi 3,4,† , Joungpyo Lim 3 , Sang-Nam Lee 5,* and Jeong-Woo Choi 3,* 1 Department of Bio & Fermentation Convergence Technology, Kookmin University, Seoul 02707, Korea; [email protected] 2 Interdisciplinary Program for Bio-health Convergence, Kookmin University, Seoul 02707, Korea 3 Department of Chemical and Biomolecular Engineering, Sogang University, Seoul 04107, Korea; [email protected] (J.-H.C.); [email protected] (J.L.) 4 School of Chemical Engineering, Jeonbuk National University, Jeonju 54896, Korea 5 Uniance Gene Inc., 1107 Teilhard Hall, 35 Baekbeom-Ro, Mapo-Gu, Seoul 04107, Korea * Correspondence: [email protected] (S.-N.L.); [email protected] (J.-W.C.); Tel.: +(82)-2-705-8480 (J.-W.C.); Fax: +(82)-2-3274-0358 (J.-W.C.) † These authors contributed equally to this work. Simple Summary: Metastasis is the main cause of cancer-related death. Circulating cancer stem cells have recently attracted attention because they have higher tumorigenicity than non-stem-like circulating tumor cells. Despite the strong scientific evidence for circulating cancer stem cells and secondary tumor formation, the exact mechanisms behind the generation and characteristics of circulating cancer stem cells are not yet fully understood because of their extreme scarcity. This Citation: Cho, H.-Y.; Choi, J.-H.; Lim, review aims to introduce the recent advances in the detection and analysis of circulating tumor cells J.; Lee, S.-N.; Choi, J.-W. Microfluidic and circulating cancer stem cells. -

Guardant Health Q4 2020 Earnings Call Operator: Ladies And
Guardant Health Q4 2020 Earnings Call Operator: Ladies and gentlemen, thank you for standing by, and welcome to the Guardant Health Q4 2020 Earnings Conference Call. At this time, all participant's lines are in a listen-only mode. After the speakers' presentation, there will a question and answer session. To ask a question during the session you will need to press star one on your telephone. Please be advised that today's conference is being recorded. If you require any further assistance, please press star zero. I would now like to hand the conference over to your speaker today, Ms. Carrie Mendivil. Thank you. Please go ahead, ma'am. Carrie Mendivil: Thank you. Earlier today, Guardant Health released financial results for the quarter and full year ended st December 31 , 2020. If you've not received this news release or if you'd like to be added to the company's distribution list, please send an e-mail to investors at guardant health dot com. Joining me today from Guardant Health is Helmy Eltoukhy, Chief Executive Officer; AmirAli Talasaz, President; and Mike Bell, Chief Financial Officer. 1 Before we begin, I'd like to remind you that management will make statements during this call that are forward-looking statements within the meaning of federal securities laws. These statements involve material risks and uncertainties that could cause actual results or events to materially differ from those anticipated. Additional information regarding these risks and uncertainties appears in the section entitled forward-looking statements in the press release Guardant issued today. For a more complete list and description, please see the Risk Factors section on the company's annual report on Form 10-K for the year st ended December 31 , 2020, and in its other filings with the Securities and Exchange Commission. -

Liquid Biopsy Cancer Biomarkers – Current Status, Future Directions
WHITE PAPER Liquid Biopsy Cancer Biomarkers – Current Status, Future Directions Liquid Biopsy: A Brief Overview Liquid biopsy offers a noninvasive or minimally invasive means of assessing physiological or pathophysiological processes that would otherwise require tissue biopsy or other invasive procedures. Most cancer diagnostic pathways currently require a fine needle aspiration at best, or an excisional biopsy at worst. Liquid biopsy, in this case, could circumvent the need for needle biopsy, endoscopy, or surgery by instead analyzing a blood sample for circulating tumor cells or circulating free nucleic acids. Since invasive diagnostic procedures are associated with a risk of hemorrhage, hematoma, tissue damage, seeding tumor cells in surrounding tissues, liquid biopsy is a safer alternative. One could also imagine using liquid biopsy to track tumor phenotype and burden over time. Physicians currently use serial CT, MRI or PET scans to assess treatment response and disease progression. These imaging studies are expensive, time-consuming, and expose patients to external radiation, intravenous contrast and/or radioactive tracers. Moreover, imaging studies do not provide any information about tumor phenotype, which can change with treatment. Today, tumor phenotype information is currently gathered through tissue biopsy. However, it is hardly ever feasible to perform serial tissue biopsies given the costs and risks to patients. Liquid biopsy, on the other hand, could provide a virtually continuous assessment of cancer burden and phenotype at relatively low cost. The patient would be at no more risk than that posed by simple blood draw. Furthermore, liquid biopsy methods may be able to detect cancer recurrence before tumor is visible on imaging or large enough for traditional tissue biopsy. -

Pre-Analytical Issues in Liquid Biopsy
J Lab Med 2020; 44(3): 117–142 Oncological Diagnostics/Liquid Profiling Edited by: C. Winter Michael Fleischhacker* and Bernd Schmidt Pre-analytical issues in liquid biopsy – where do we stand? https://doi.org/10.1515/labmed-2019-0167 The analysis of cell-free nucleic acids (which has mis- Received October 1, 2019; accepted February 26, 2020; previously leadingly been coined liquid biopsy, more precisely would published online April 7, 2020 be “liquid profiling”) may be useful in different areas, for the diagnosis of patients with different diseases and espe- Abstract: It is well documented that in the chain from sam- cially for cancer patients, treatment monitoring, tracking ple to the result in a clinical laboratory, the pre-analytical of minimal residual disease (early appearance of treat- phase is the weakest and most vulnerable link. This also ment resistance), and for the establishment of the prog- holds for the use and analysis of extracellular nucleic nosis for these patients. While the latter points might be acids. In this short review, we will summarize and criti- within tangible reach, its usefulness as a diagnostic tool cally evaluate the most important steps of the pre-analyt- is still heavily debated. The reasons for this include, but ical phase, i.e. the choice of the best control population are not limited to: for the patients to be analyzed, the actual blood draw, the – the very low concentration of DNA with tumor-asso- choice of tubes for blood drawing, the impact of delayed ciated alterations in the blood/body fluids of early- processing of blood samples, the best method for getting stage cancer patients; rid of cells and debris, the choice of matrix, i.e.