Investigating Cell-Free DNA in Liquid Biopsy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Absolute Measurement of the Tissue Origins of Cell-Free DNA in the Healthy State and Following Paracetamol Overdose Danny Laurent1, Fiona Semple1, Philip J
Laurent et al. BMC Medical Genomics (2020) 13:60 https://doi.org/10.1186/s12920-020-0705-2 RESEARCH ARTICLE Open Access Absolute measurement of the tissue origins of cell-free DNA in the healthy state and following paracetamol overdose Danny Laurent1, Fiona Semple1, Philip J. Starkey Lewis2, Elaine Rose1, Holly A. Black1, Jennifer Coe1, Stuart J. Forbes2, Mark J. Arends3, James W. Dear4 and Timothy J. Aitman1* Abstract Background: Despite the emergence of cell-free DNA (cfDNA) as a clinical biomarker in cancer, the tissue origins of cfDNA in healthy individuals have to date been inferred only by indirect and relative measurement methods, such as tissue-specific methylation and nucleosomal profiling. Methods: We performed the first direct, absolute measurement of the tissue origins of cfDNA, using tissue-specific knockout mouse strains, in both healthy mice and following paracetamol (APAP) overdose. We then investigated the utility of total cfDNA and the percentage of liver-specific cfDNA as clinical biomarkers in patients presenting with APAP overdose. Results: Analysis of cfDNA from healthy tissue-specific knockout mice showed that cfDNA originates predominantly from white and red blood cell lineages, with minor contribution from hepatocytes, and no detectable contribution from skeletal and cardiac muscle. Following APAP overdose in mice, total plasma cfDNA and the percentage fraction originating from hepatocytes increased by ~ 100 and ~ 19-fold respectively. Total cfDNA increased by an average of more than 236-fold in clinical samples from APAP overdose patients with biochemical evidence of liver injury, and 18-fold in patients without biochemically apparent liver injury. Measurement of liver-specific cfDNA, using droplet digital PCR and methylation analysis, revealed that the contribution of liver to cfDNA was increased by an average of 175-fold in APAP overdose patients with biochemically apparent liver injury compared to healthy subjects, but was not increased in overdose patients with normal liver function tests. -

Health Sciences Center
Faculty of Allied Health Sciences Handbook: 2020-2021 KUWAIT UNIVERSITY HEALTH SCIENCES CENTRE 1 | P a g e KUWAIT UNIVERSITY HEALTH SCIENCES CENTRE FACULTY OF ALLIED HEALTH SCIENCES Established: 1982 HANDBOOK 2020-2021 2 | P a g e Department of MEDICAL LABORATORY SCIENCES [MLS] 3 | P a g e DEPARTMENT OF MEDICAL LABORATORY SCIENCES Medical Laboratory Sciences offers opportunities for those interested in biological and chemical sciences, leading to a career in the health service or in research. Medical laboratory scientists are professionals who perform laboratory tests and analyses that assist physicians in the diagnosis and treatment of patients. They also assist in research and the development of new laboratory tests. The various studies include chemical and physical analysis of body fluids (clinical chemistry and urinalysis); examination of blood and its component cells (haematology); isolation and identification of bacteria, fungi, viruses and parasites (clinical microbiology and parasitology); testing of blood serum for antibodies indicative of specific diseases (immunology and serology) and collection, storage of blood, pretransfusion testing and other immunohaematological procedures (blood banking). In addition, medical laboratory scientists prepare tissues for histopathological, cytological and cytogenetic examination. They must know the theory and scientific fundamentals as well as the procedures for testing. Medical laboratory scientists work in hospital clinical laboratories, medical schools, research institutions, public health agencies and related organizations. MISSION AND OBJECTIVES Mission The mission of the Department of Medical Laboratory Sciences is to educate and train skillful, knowledgeable and committed Medical Laboratory Scientists who have breadth of knowledge and competence in the various aspects of Medical Laboratory Sciences, who shall adhere to professional ethics, and who can contribute successfully as Medical Laboratory Scientists in the health care team. -

797 Circulating Tumor DNA and Circulating Tumor Cells for Cancer
Medical Policy Circulating Tumor DNA and Circulating Tumor Cells for Cancer Management (Liquid Biopsy) Table of Contents • Policy: Commercial • Coding Information • Information Pertaining to All Policies • Policy: Medicare • Description • References • Authorization Information • Policy History • Endnotes Policy Number: 797 BCBSA Reference Number: 2.04.141 Related Policies Biomarkers for the Diagnosis and Cancer Risk Assessment of Prostate Cancer, #336 Policy1 Commercial Members: Managed Care (HMO and POS), PPO, and Indemnity Plasma-based comprehensive somatic genomic profiling testing (CGP) using Guardant360® for patients with Stage IIIB/IV non-small cell lung cancer (NSCLC) is considered MEDICALLY NECESSARY when the following criteria have been met: Diagnosis: • When tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP or invasive biopsy is medically contraindicated), AND • When prior results for ALL of the following tests are not available: o EGFR single nucleotide variants (SNVs) and insertions and deletions (indels) o ALK and ROS1 rearrangements o PDL1 expression. Progression: • Patients progressing on or after chemotherapy or immunotherapy who have never been tested for EGFR SNVs and indels, and ALK and ROS1 rearrangements, and for whom tissue-based CGP is infeasible (i.e., quantity not sufficient for tissue-based CGP), OR • For patients progressing on EGFR tyrosine kinase inhibitors (TKIs). If no genetic alteration is detected by Guardant360®, or if circulating tumor DNA (ctDNA) is insufficient/not detected, tissue-based genotyping should be considered. Other plasma-based CGP tests are considered INVESTIGATIONAL. CGP and the use of circulating tumor DNA is considered INVESTIGATIONAL for all other indications. 1 The use of circulating tumor cells is considered INVESTIGATIONAL for all indications. -

MLH1 Single-Nucleotide Variant in Circulating Tumor DNA
www.nature.com/scientificreports OPEN MLH1 single‑nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma Soon Sun Kim1,4, Jung Woo Eun1,4, Ji‑Hye Choi2, Hyun Goo Woo2, Hyo Jung Cho1, Hye Ri Ahn3, Chul Won Suh3, Geum Ok Baek1, Sung Won Cho1 & Jae Youn Cheong1* Liquid biopsy can provide a strong basis for precision medicine. We aimed to identify novel single‑ nucleotide variants (SNVs) in circulating tumor DNA (ctDNA) in patients with hepatocellular carcinoma (HCC). Deep sequencing of plasma‑derived ctDNA from 59 patients with HCC was performed using a panel of 2924 SNVs in 69 genes. In 55.9% of the patients, at least one somatic mutation was detected. Among 25 SNVs in 12 genes, four frequently observed SNVs, MLH1 (13%), STK11 (13%), PTEN (9%), and CTNNB1 (4%), were validated using droplet digital polymerase chain reaction with ctDNA from 62 patients with HCC. Three candidate SNVs were detected in 35.5% of the patients, with a frequency of 19% for MLH1 chr3:37025749T>A, 11% for STK11 chr19:1223126C>G, and 8% for PTEN chr10:87864461C>G. The MLH1 and STK11 SNVs were also confrmed in HCC tissues. The presence of the MLH1 SNV, in combination with an increased ctDNA level, predicted poor overall survival among 107 patients. MLH1 chr3:37025749T>A SNV detection in ctDNA is feasible, and thus, ctDNA can be used to detect somatic mutations in HCC. Furthermore, the presence or absence of the MLH1 SNV in ctDNA, combined with the ctDNA level, can predict the prognosis of patients with HCC. -

Brain – Necrosis
Brain – Necrosis 1 Brain – Necrosis 2 Brain – Necrosis Figure Legend: Figure 1 Appearance of a thalamic infarct at low magnification, identified by pallor within the zone of the black arrows, in an F344/N rat. The dentate gyrus of the hippocampus is identified by a white arrow. This infarct was the result of an arterial embolus (arrowhead), shown at higher magnification in Figure 2. Figure 2 Arterial embolus from Figure 1 at higher magnification, in an F344/N rat. Figure 3 Acute necrosis of the posterior colliculus, a bilaterally symmetrical lesion (arrows), in the whole mount of a section in a male F344/N rat from an acute study. This resulted from the selective vulnerability of this brain region to toxin- induced impaired energy metabolism. The arrowhead identifies necrosis of the nucleus of the lateral lemniscus. Figure 4 Similar regionally selective bilateral brain necrosis of the parietal cortex area 1 (blue arrow), thalamus (arrowhead), and retrosplenial cortex (white arrow) in a treated male F344/N rat from an acute study, all resulting from the same toxic compound as used in Figure 3. Figure 5 Unusual form of malacia (total regional necrosis) of the spinal cord in the dorsal spinal funiculi (arrow) in a female F344/N rat from a chronic study. Figure 6 A cortical infarct with gliosis and capillary hyperplasia (arrow) from a male B6C3F1 mouse in a chronic study. Figure 7 A more advanced stage of cortical infarction (arrows) in a treated female B6C3F1 mouse from a chronic chronic inhalation study. Figure 8 Morphology of an infarct of known duration (arrow) in an F344/N rat with experimental infarction. -
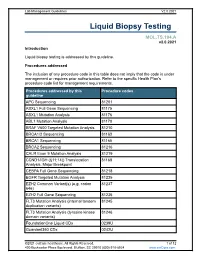
Liquid Biopsy Testing
Lab Management Guidelines V2.0.2021 Liquid Biopsy Testing MOL.TS.194.A v2.0.2021 Introduction Liquid biopsy testing is addressed by this guideline. Procedures addressed The inclusion of any procedure code in this table does not imply that the code is under management or requires prior authorization. Refer to the specific Health Plan's procedure code list for management requirements. Procedures addressed by this Procedure codes guideline APC Sequencing 81201 ASXL1 Full Gene Sequencing 81175 ASXL1 Mutation Analysis 81176 ABL1 Mutation Analysis 81170 BRAF V600 Targeted Mutation Analysis 81210 BRCA1/2 Sequencing 81163 BRCA1 Sequencing 81165 BRCA2 Sequencing 81216 CALR Exon 9 Mutation Analysis 81219 CCND1/IGH (t(11;14)) Translocation 81168 Analysis, Major Breakpoint CEBPA Full Gene Sequencing 81218 EGFR Targeted Mutation Analysis 81235 EZH2 Common Variant(s) (e.g. codon 81237 646) EZH2 Full Gene Sequencing 81236 FLT3 Mutation Analysis (internal tandem 81245 duplication variants) FLT3 Mutation Analysis (tyrosine kinase 81246 domain variants) FoundationOne Liquid CDx 0239U Guardant360 CDx 0242U ©2021 eviCore healthcare. All Rights Reserved. 1 of 12 400 Buckwalter Place Boulevard, Bluffton, SC 29910 (800) 918-8924 www.eviCore.com Lab Management Guidelines V2.0.2021 Procedures addressed by this Procedure codes guideline Hematolymphoid Neoplasm Molecular 81450 Profiling; 5-50 genes IDH1 Mutation Analysis 81120 IDH2 Mutation Analysis 81121 IGH@/BCL2 (t(14;18)) Translocation 81278 Analysis, Major Breakpoint Region (MBR) and Minor Cluster Region (mcr) Breakpoints JAK2 Targeted Mutation Analysis (e.g 81279 exons 12 and 13) JAK2 V617F Targeted Mutation Analysis 81270 KIT Targeted Sequence Analysis 81272 KIT D816 Targeted Mutation Analysis 81273 KRAS Exon 2 Targeted Mutation Analysis 81275 KRAS Targeted Mutation Analysis, 81276 Additional Variants MGMT Promoter Methylation Analysis 81287 MLH1 Sequencing 81292 Molecular Tumor Marker Test 81400 81401 81402 g 81403 n i 81405 t 81406 s e T 81407 81408 y s 81479 p o Molecular Tumor Marker Test 88271 i B MPL Common Variants (e.g. -

Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas
Published OnlineFirst June 24, 2015; DOI: 10.1158/2159-8290.CD-15-0274 ReseaRch BRief Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas Oliver A. Zill1,2,3, Claire Greene2,4, Dragan Sebisanovic3, Lai Mun Siew3, Jim Leng2,4, Mary Vu5, Andrew E. Hendifar6, Zhen Wang2, Chloe E. Atreya2,4, Robin K. Kelley2,4, Katherine Van Loon2,4, Andrew H. Ko2,4, Margaret A. Tempero2,4, Trever G. Bivona2,4, Pamela N. Munster2,4, AmirAli Talasaz3, and Eric A. Collisson2,4 AbstRact Patients with pancreatic and biliary carcinomas lack personalized treatment options, in part because biopsies are often inadequate for molecular characteriza- tion. Cell-free DNA (cfDNA) sequencing may enable a precision oncology approach in this setting. We attempted to prospectively analyze 54 genes in tumor and cfDNA for 26 patients. Tumor sequencing failed in 9 patients (35%). In the remaining 17, 90.3% (95% confidence interval, 73.1%–97.5%) of mutations detected in tumor biopsies were also detected in cfDNA. The diagnostic accuracy of cfDNA sequencing was 97.7%, with 92.3% average sensitivity and 100% specificity across five informative genes. Changes in cfDNA correlated well with tumor marker dynamics in serial sampling (r = 0.93). We demonstrate that cfDNA sequencing is feasible, accurate, and sensitive in identifying tumor-derived mutations without prior knowledge of tumor genotype or the abundance of circulating tumor DNA. cfDNA sequencing should be considered in pancreatobiliary cancer trials where tissue sampling is unsafe, infeasible, or otherwise unsuccessful. SIGNIFICANCE: Precision medicine efforts in biliary and pancreatic cancers have been frustrated by difficulties in obtaining adequate tumor tissue for next-generation sequencing. -

Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine Giovanna Rossi1 and Michail Ignatiadis2
Published OnlineFirst May 20, 2019; DOI: 10.1158/0008-5472.CAN-18-3402 Cancer Review Research Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine Giovanna Rossi1 and Michail Ignatiadis2 Abstract New sensitive assays are currently available for the detec- Multiple studies are underway to assess the clinical utility of tion of circulating tumor DNA (ctDNA) and circulating tumor CTC and ctDNA in different settings (treatment-na€ve vs. cells (CTC). However, there remains a need for standardiza- resistant, adjuvant vs. metastatic) and for different treatment tion of preanalytical issues and cross-platform comparison modalities (systemic therapy, surgery, radiation therapy). studies. Liquid biopsies are being evaluated for treatment This review aims to map the challenges that remain to be selection, for monitoring disease response and resistance, for addressed before liquid biopsies can be widely used for cancer tracking minimal residual disease, and for cancer diagnosis. management. Introduction applications of CTCs and ctDNA that are currently being explored are summarized in Fig. 1A. In the era of precision medicine, liquid biopsies are increasingly being studied as a tool that can capture tumor evolution in real time and thus guide systemic treatment. In this article, we will refer to the analysis of circulating tumor DNA (ctDNA) and circulating Challenges Associated with Preanalytical tumor cells (CTC) only and we will not cover other liquid biopsy Issues and the Analytical Validity of Liquid biomarkers such as circulating RNAs, proteins, metabolites, and Biopsy Assays exosomes. For CTC and ctDNA assays, there is a need to standardize Sampling a patient's blood may give information about the preanalytical variables and for cross-platform comparison stud- genomic profile of a given cancer (1–3) and provide an assess- ies. -

Liquid Biopsy Analysis in Clinical Practice: Focus on Lung Cancer
Review Liquid Biopsy Analysis in Clinical Practice: Focus on Lung Cancer Pasquale Pisapia 1 , Francesco Pepe 1, Antonino Iaccarino 1, Roberta Sgariglia 1, Mariantonia Nacchio 1, Gianluca Russo 1 , Gianluca Gragnano 1, Elalah Mosaieby 2, Giancarlo Troncone 1,* and Umberto Malapelle 1 1 Department of Public Health, University of Naples Federico II, 80131 Naples, Italy; [email protected] (P.P.); [email protected] (F.P.); [email protected] (A.I.); [email protected] (R.S.); [email protected] (M.N.); [email protected] (G.R.); [email protected] (G.G.); [email protected] (U.M.) 2 Department of Cellular and Molecular Biology, University of Mazandaran, Mazandaran 48175-866, Iran; [email protected] * Correspondence: [email protected] Abstract: Lung cancer is the leading cause of cancer death worldwide. Despite the emergence of highly effective targeted therapies, up to 30% of advanced stage non-small cell lung cancer (NSCLC) patients do not undergo tissue molecular testing because of scarce tissue availability. Liquid biopsy, on the other hand, offers these patients a valuable opportunity to receive the best treatment options in a timely manner. Indeed, besides being much faster and less invasive than conventional tissue- based analysis, it can also yield specific information about the genetic make-up and evolution of patients’ tumors. However, several issues, including lack of standardized protocols for sample collection, processing, and interpretation, still need to be addressed before liquid biopsy can be Citation: Pisapia, P.; Pepe, F.; fully incorporated into routine oncology practice. Here, we reviewed the most important challenges Iaccarino, A.; Sgariglia, R.; Nacchio, hindering the implementation of liquid biopsy in oncology practice, as well as the great advantages M.; Russo, G.; Gragnano, G.; Mosaieby, E.; Troncone, G.; Malapelle, of this approach for the treatment of NSCLC patients. -

Donor-Derived Cell-Free DNA in Kidney Transplantation As a Potential Rejection Biomarker: a Systematic Literature Review
Journal of Clinical Medicine Review Donor-Derived Cell-Free DNA in Kidney Transplantation as a Potential Rejection Biomarker: A Systematic Literature Review Adrian Martuszewski 1 , Patrycja Paluszkiewicz 1 , Magdalena Król 2, Mirosław Banasik 1 and Marta Kepinska 3,* 1 Department of Nephrology and Transplantation Medicine, Wroclaw Medical University, Borowska 213, 50-556 Wroclaw, Poland; [email protected] (A.M.); [email protected] (P.P.); [email protected] (M.B.) 2 Students Scientific Association, Department of Biomedical and Environmental Analysis, Faculty of Pharmacy, Wroclaw Medical University, 50-556 Wroclaw, Poland; [email protected] 3 Department of Biomedical and Environmental Analyses, Faculty of Pharmacy, Wroclaw Medical University, Borowska 211, 50-556 Wroclaw, Poland * Correspondence: [email protected]; Tel.: +48-71-784-0171 Abstract: Kidney transplantation (KTx) is the best treatment method for end-stage kidney disease. KTx improves the patient’s quality of life and prolongs their survival time; however, not all patients benefit fully from the transplantation procedure. For some patients, a problem is the premature loss of graft function due to immunological or non-immunological factors. Circulating cell-free DNA (cfDNA) is degraded deoxyribonucleic acid fragments that are released into the blood and other body fluids. Donor-derived cell-free DNA (dd-cfDNA) is cfDNA that is exogenous to the patient and comes from a transplanted organ. As opposed to an invasive biopsy, dd-cfDNA can be detected by a non-invasive analysis of a sample. The increase in dd-cfDNA concentration occurs even before the creatinine level starts rising, which may enable early diagnosis of transplant injury and adequate Citation: Martuszewski, A.; treatment to avoid premature graft loss. -

Circulating Tumor DNA As an Emerging Liquid Biopsy Biomarker
Int. J. Biol. Sci. 2020, Vol. 16 1551 Ivyspring International Publisher International Journal of Biological Sciences 2020; 16(9): 1551-1562. doi: 10.7150/ijbs.44024 Review Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma Xiaolin Wu1, Jiahui Li1, Asmae Gassa2, Denise Buchner1, Hakan Alakus1, Qiongzhu Dong3, Ning Ren4, Ming Liu5, Margarete Odenthal6, Dirk Stippel1, Christiane Bruns1, Yue Zhao1,7, and Roger Wahba1 1. Department of General, Visceral, Cancer and Transplantation Surgery, University Hospital of Cologne, Kerpener Straße 62, 50937, Cologne, Germany. 2. Department of Cardiothoracic Surgery, Heart Center, University Hospital of Cologne, Germany, Kerpener Straße 62, 5.937 Cologne, Germany. 3. Department of General Surgery, Huashan Hospital & Cancer Metastasis Institute & Institutes of Biomedical Sciences, Fudan University, 200032, Shanghai, P.R. China. 4. Liver Cancer Institute & Zhongshan Hospital; Department of Surgery, Institute of Fudan-Minhang Academic Health System, Minhang Branch, Zhongshan Hospital, Fudan University, 200032, Shanghai, P.R. China. 5. Affiliated Cancer Hospital & Institute of Guangzhou Medical University; Key Laboratory of Protein Modification and Degradation, School of Basic Medical Sciences, 510095, Guangzhou, P.R. China. 6. Institute of Pathology, University Hospital of Cologne, 50937, Cologne, Germany. 7. Department of General, Visceral und Vascular Surgery, Otto-von-Guericke University, 39120, Magdeburg, Germany. Corresponding authors: Roger Wahba ([email protected]; Tel: +49-221-478-4803); Yue Zhao (yue.zhao @uk-koeln.de; Tel: +49-221-478-30601). © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). -

Role of Histopathological Examination in Medicolegal Autopsies in Unravelling Pathology Section Precise Causes of Mortality
DOI: 10.7860/NJLM/2021/48233:2522 Original Article Role of Histopathological Examination in Medicolegal Autopsies in Unravelling Pathology Section Precise Causes of Mortality DIVYA SHARMA1, ANSHU GUPTA2, KHUSHBOO DEWAN3, KARSING PATIRI4, KUSUM GUPTA5, USHA RANI SINGH6 ABSTRACT Histopathological examination was performed in 96 cases out of Introduction: Medicolegal autopsies are performed to determine which 10 were excluded due to autolysis (n=86). Haemotoxilin the cause and manner of death. Histopathological examination and Eosin (H&E)-stained slides were examined and special is reserved for only those cases where Cause Of Death (COD) is stains and Immunohistochemistry (IHC) applied wherever not readily apparent on autopsy. However, there are conflicting required. Gross and histopathological findings were recorded views regarding the utility of histopathological examination in along with autopsy findings and clinical history. The results were medicolegal cases. tabulated and statistical analysis was done using the Chi-square and Fischer’s test to look for any significance and association Aim: To examine the role of histopathological examination in between gross and microscopic findings in various organs. The unravelling specific causes of mortality in two settings: 1) where p-value of <0.05 was considered significant. collaborative clinical history and gross autopsy findings were available; 2) where definitive cause could be discovered only Results: Histopathological examination was conclusive in at the time of microscopic examination, thus altering its legal ascertaining the specific COD in 30/86 cases (35%). These were implications. categorised as pulmonary causes (27) including one case each Materials and Methods: This was a retrospective observational of fat embolism and Amniotic Fluid Embolism (AFE) and cardiac study including all medicolegal autopsy cases, in which causes (3).