Diagnosis of Lethal Or Prenatal-Onset Autosomal Recessive Disorders by Parental Exome Sequencing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Crouzon Syndrome with Ophthalmological Complications
Journal of Rawalpindi Medical College (JRMC); 2012;16(1):80-81 Case Report Crouzon Syndrome With Ophthalmological Complications Rahela Nasir Paediatric Department, Capital Hospital, CDA Islamabad. Crouzon Syndrome is characterized by premature bilateral optic atrophy. Other facial features included a craniosynostosis. It has an autosomal dominant inheritance prominent nose and deep and narrow palate. No but represents fresh mutation also. Other craniofacial digital abnormalities were seen. No dental aplasia was abnormalities include ocular proptosis caused by shallow present. Systemic examination revealed no orbits with or without divergent strabismus. There may be abnormality. increased intracranial pressure for which surgical morcellation procedures are indicated. A case of He was diagnosed as a case of Crouzon syndrome craniosynostosis is reported which is diagnosed as Crouzon with ocular complications on clinical basis. He was Syndrome with ocular complications on clinical grounds. investigated for microcephaly and suspected Split craniotomy was performed by a neurosurgeon to craniosynostosis. Radiographs of skull showed small relieve raised intracranial pressure and to enhance brain sized skull with early closure of sutures and fontanelle growth. Crouzon Syndrome was originally described in 1912 suggestive of craniosynstosis(Fig 2). A hammered- by Crouzon in a mother and her daughter. It is an autosomal silver (beaten metal/ copper beaten) appearance was dominant inherited disorder but represents fresh mutation also seen due to raised intracranial pressure and also. Crouzon syndrome is characterized by premature compression of the developing brain on the fused craniosynostosis which is quite variable but the coronal suture is nearly always bilaterally involved. Craniofacial bone. CT scan brain with contrast was done which abnormalities include brachycephaly, shallow orbits and confirmed the features suggestive of craniosynostosis maxillary hypoplasia. -
Macrocephaly Information Sheet 6-13-19
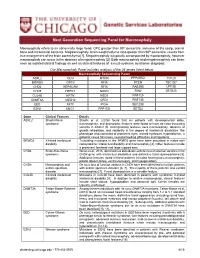
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Sotos Syndrome
European Journal of Human Genetics (2007) 15, 264–271 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg PRACTICAL GENETICS In association with Sotos syndrome Sotos syndrome is an autosomal dominant condition characterised by a distinctive facial appearance, learning disability and overgrowth resulting in tall stature and macrocephaly. In 2002, Sotos syndrome was shown to be caused by mutations and deletions of NSD1, which encodes a histone methyltransferase implicated in chromatin regulation. More recently, the NSD1 mutational spectrum has been defined, the phenotype of Sotos syndrome clarified and diagnostic and management guidelines developed. Introduction In brief Sotos syndrome was first described in 1964 by Juan Sotos Sotos syndrome is characterised by a distinctive facial and the major diagnostic criteria of a distinctive facial appearance, learning disability and childhood over- appearance, childhood overgrowth and learning disability growth. were established in 1994 by Cole and Hughes.1,2 In 2002, Sotos syndrome is associated with cardiac anomalies, cloning of the breakpoints of a de novo t(5;8)(q35;q24.1) renal anomalies, seizures and/or scoliosis in B25% of translocation in a child with Sotos syndrome led to the cases and a broad variety of additional features occur discovery that Sotos syndrome is caused by haploinsuffi- less frequently. ciency of the Nuclear receptor Set Domain containing NSD1 abnormalities, such as truncating mutations, protein 1 gene, NSD1.3 Subsequently, extensive analyses of missense mutations in functional domains, partial overgrowth cases have shown that intragenic NSD1 muta- gene deletions and 5q35 microdeletions encompass- tions and 5q35 microdeletions encompassing NSD1 cause ing NSD1, are identifiable in the majority (490%) of 490% of Sotos syndrome cases.4–10 In addition, NSD1 Sotos syndrome cases. -

KBG Syndrome Presenting with Brachydactyly Type E
Bone 123 (2019) 18–22 Contents lists available at ScienceDirect Bone journal homepage: www.elsevier.com/locate/bone Case Report KBG syndrome presenting with brachydactyly type E T ⁎ Renata Libiantoa,b,c, , Kathy HC Wud,e,f,g, Sophie Deveryd, John A Eismana,b,f,h, Jackie R Centera,b,f,h a Bone Division, Garvan Institute of Medical Research, Sydney, Australia b Department of Endocrinology, St Vincent's Hospital Sydney, Australia c Department of Medicine, The University of Melbourne, Australia d Clinical Genomics Unit, St Vincent's Hospital Sydney, Australia e Discipline of Genetic Medicine, University of Sydney, Australia f School of Medicine, UNSW, Sydney, Australia g Genomics and Epigenetics Division, Garvan Institute of Medical Research, Sydney, Australia h School of Medicine Sydney, University of Notre Dame, Australia ARTICLE INFO ABSTRACT Keywords: We report the case of a young woman who presented at age 10 years with height on the tenth centile, bra- KBG syndrome chydactyly type E and mild developmental delay. Biochemistry and hormonal profiles were normal. Differential ANKRD11 gene diagnoses considered included Albright hereditary osteodystrophy without hormone resistance (a.k.a pseu- Skeletal disorder dopseudohypoparathyroidism), 2q37 microdeletion syndrome and acrodysostosis. She had a normal karyotype Brachydactyly type E and normal FISH of 2q37. Whole genome sequencing (WGS) identified a mutation in the ANKRD11 gene as- sociated with KBG syndrome. We review the clinical features of the genetic syndromes considered, and suggest KBG syndrome be considered in patients presenting with syndromic brachydactyly type E, especially if short stature and developmental delay are also present. 1. Case support in a mainstream school for specific learning difficulties. -

Accelerating Matchmaking of Novel Dysmorphology Syndromes Through Clinical and Genomic Characterization of a Large Cohort
ORIGINAL RESEARCH ARTICLE © American College of Medical Genetics and Genomics Accelerating matchmaking of novel dysmorphology syndromes through clinical and genomic characterization of a large cohort Ranad Shaheen, PhD1, Nisha Patel, PhD1, Hanan Shamseldin, MSc1, Fatema Alzahrani, BSc1, Ruah Al-Yamany, MD1, Agaadir ALMoisheer, MSc1, Nour Ewida, BSc1, Shamsa Anazi, MSc1, Maha Alnemer, MD2, Mohamed Elsheikh, MD3, Khaled Alfaleh, MD3,4, Muneera Alshammari, MD4, Amal Alhashem, MD5, Abdullah A. Alangari, MD4, Mustafa A. Salih, MD4, Martin Kircher, MD6, Riza M. Daza, PhD6, Niema Ibrahim, BSc1, Salma M. Wakil, PhD1, Ahmed Alaqeel, MD7, Ikhlas Altowaijri, MD7, Jay Shendure, MD, PhD6, Amro Al-Habib, MD7, Eissa Faqieh, MD8 and Fowzan S. Alkuraya, MD1,9 Purpose: Dysmorphology syndromes are among the most common and C3ORF17). A significant minority of the phenotypes (6/31, 19%), referrals to clinical genetics specialists. Inability to match the dysmor- however, were caused by genes known to cause Mendelian pheno- phology pattern to a known syndrome can pose a major diagnostic chal- types, thus expanding the phenotypic spectrum of the diseases linked lenge. With an aim to accelerate the establishment of new syndromes to these genes. The conspicuous inheritance pattern and the highly and their genetic etiology, we describe our experience with multiplex specific phenotypes appear to have contributed to the high yield consanguineous families that appeared to represent novel autosomal (90%) of plausible molecular diagnoses in our study cohort. recessive dysmorphology syndromes at the time of evaluation. Conclusion: Reporting detailed clinical and genomic analysis of Methods: Combined autozygome/exome analysis of multiplex a large series of apparently novel dysmorphology syndromes will consanguineous families with apparently novel dysmorphology syn- likely lead to a trend to accelerate the establishment of novel syn- dromes. -

Lieshout Van Lieshout, M.J.S
EXPLORING ROBIN SEQUENCE Manouk van Lieshout Van Lieshout, M.J.S. ‘Exploring Robin Sequence’ Cover design: Iliana Boshoven-Gkini - www.agilecolor.com Thesis layout and printing by: Ridderprint BV - www.ridderprint.nl ISBN: 978-94-6299-693-9 Printing of this thesis has been financially supported by the Erasmus University Rotterdam. Copyright © M.J.S. van Lieshout, 2017, Rotterdam, the Netherlands All rights reserved. No parts of this thesis may be reproduced, stored in a retrieval system, or transmitted in any form or by any means without permission of the author or when appropriate, the corresponding journals Exploring Robin Sequence Verkenning van Robin Sequentie Proefschrift ter verkrijging van de graad van doctor aan de Erasmus Universiteit Rotterdam op gezag van de rector magnificus Prof.dr. H.A.P. Pols en volgens besluit van het College voor Promoties. De openbare verdediging zal plaatsvinden op woensdag 20 september 2017 om 09.30 uur door Manouk Ji Sook van Lieshout geboren te Seoul, Korea PROMOTIECOMMISSIE Promotoren: Prof.dr. E.B. Wolvius Prof.dr. I.M.J. Mathijssen Overige leden: Prof.dr. J.de Lange Prof.dr. M. De Hoog Prof.dr. R.J. Baatenburg de Jong Copromotoren: Dr. K.F.M. Joosten Dr. M.J. Koudstaal TABLE OF CONTENTS INTRODUCTION Chapter I: General introduction 9 Chapter II: Robin Sequence, A European survey on current 37 practice patterns Chapter III: Non-surgical and surgical interventions for airway 55 obstruction in children with Robin Sequence AIRWAY OBSTRUCTION Chapter IV: Unravelling Robin Sequence: Considerations 79 of diagnosis and treatment Chapter V: Management and outcomes of obstructive sleep 95 apnea in children with Robin Sequence, a cross-sectional study Chapter VI: Respiratory distress following palatal closure 111 in children with Robin Sequence QUALITY OF LIFE Chapter VII: Quality of life in children with Robin Sequence 129 GENERAL DISCUSSION AND SUMMARY Chapter VIII: General discussion 149 Chapter IX: Summary / Nederlandse samenvatting 169 APPENDICES About the author 181 List of publications 183 Ph.D. -

Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S
CLINICAL REPORT Guidance for the Clinician in Rendering Pediatric Care Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S. Dias, MD, FAAP, FAANS,a Thomas Samson, MD, FAAP,b Elias B. Rizk, MD, FAAP, FAANS,a Lance S. Governale, MD, FAAP, FAANS,c Joan T. Richtsmeier, PhD,d SECTION ON NEUROLOGIC SURGERY, SECTION ON PLASTIC AND RECONSTRUCTIVE SURGERY Pediatric care providers, pediatricians, pediatric subspecialty physicians, and abstract other health care providers should be able to recognize children with abnormal head shapes that occur as a result of both synostotic and aSection of Pediatric Neurosurgery, Department of Neurosurgery and deformational processes. The purpose of this clinical report is to review the bDivision of Plastic Surgery, Department of Surgery, College of characteristic head shape changes, as well as secondary craniofacial Medicine and dDepartment of Anthropology, College of the Liberal Arts characteristics, that occur in the setting of the various primary and Huck Institutes of the Life Sciences, Pennsylvania State University, State College, Pennsylvania; and cLillian S. Wells Department of craniosynostoses and deformations. As an introduction, the physiology and Neurosurgery, College of Medicine, University of Florida, Gainesville, genetics of skull growth as well as the pathophysiology underlying Florida craniosynostosis are reviewed. This is followed by a description of each type of Clinical reports from the American Academy of Pediatrics benefit from primary craniosynostosis (metopic, unicoronal, bicoronal, sagittal, lambdoid, expertise and resources of liaisons and internal (AAP) and external reviewers. However, clinical reports from the American Academy of and frontosphenoidal) and their resultant head shape changes, with an Pediatrics may not reflect the views of the liaisons or the emphasis on differentiating conditions that require surgical correction from organizations or government agencies that they represent. -

Bagatelle Cassidy Syndrome: Macrocephaly
Journal of Clinical and Experimental Genetics Volume 1 | Issue 1 Case Report Open Access Bagatelle Cassidy Syndrome: Macrocephaly with Fronto-Facial Dysmorphism and Hearing Loss; an Extremely Rare Case Avina Fierro JA*1 and Hernandez Avina DA2 1Department of Pediatric Dysmorphology, IMSS Medical Center; Guadalajara, Mexico 2General Hospital Zona 2, IMSS Aguascalientes, Mexico *Corresponding author: Avina Fierro JA, Department of Pediatric Dysmorphology, IMSS Medical Center; Guadalajara, Mexico, Tel: (52)3336-743701, E-mail: [email protected] Citation: Avina Fierro JA, Hernandez Avina DA (2015) Bagatelle Cassidy Syndrome: Macrocephaly with Fronto-Facial Dysmorphism and Hearing Loss; an Extremely Rare Case. J Clin Exp Gen 1(1): 101 Abstract Bagatelle Cassidy syndrome is a very rare disease first described in 1995 in a boy with macrocephaly, hypertelorism, hearing loss, developmental delay and facial dysmorphism. The cause of this syndrome is unknown and has no specific diagnostic test. We report the case of a 9-year-old girl with a cerebro-fronto-facial malformation syndrome with macrocephaly, with significant decrease of the hearing, moderate hearing loss (50 db HL at 4000 Hz); mild mental retardation (IQ 50) and noticeable speech delay, psychomotor and developmental delay. Facial anomalies show prominent forehead (frontal bossing) , hypertelorism, downslanting palpebral fissures, flat nasal bridge, short nose with anteverted nostrils; broad columella, long smooth philtrum and carp-like mouth; this phenotype represents the specific -

Treacher Collins Syndrome: a Case Report and Review of Literature Priya Thomas1, Rekha Krishnapillai2, Bindhu P Ramakrishnan3
CASE REPORT Treacher Collins Syndrome: A Case Report and Review of Literature Priya Thomas1 , Rekha Krishnapillai2 , Bindhu P Ramakrishnan3 ABSTRACT Introduction: Treacher Collins syndrome (TCS) is a rare autosomal dominant disorder of craniofacial morphogenesis. The congenital malformation is restricted to first and second branchial arches which affects the mandible, maxillozygomatic complex, ears, and eyelids. Many cases also manifest with dental anomalies resultant of the craniofacial abnormality. The severity of the facial deformity varies with affected individuals based on the variability of penetrance. This paper reports a case of a 31-year-old male with TCS. Case description: A 31-year-old male patient reported to the institution with a chief complaint of forwardly placed upper front teeth since 15 years. He also reported difficulty in speech, loosened anterior teeth since 5 years, and mild pain on gums while having food and during speech. The patient also gave a history of surgical correction for the “widened” mouth at the age of 2 years and has been on medications for recurrent ear infections. Management: Treatment consists of surgical management of anomalous facial structures. Preoperative planning and evaluation should begin as early as possible. A staged treatment and multidisciplinary craniofacial team is required to coordinate cranio-orofacial, dental, ocular, and pediatric care beginning in the early neonatal life throughout childhood. Conclusion: Prenatal diagnosis and genetic counseling are mandatory to help parents to make decisions regarding pregnancy. These patients require care from birth through adulthood. Proper treatment planning, counseling, and surgical management are essential for optimizing patient outcomes. Keywords: Autosomal dominance, Craniofacial complex, Hypoplasia, Treacher Collins syndrome. -

Association of Generalized Aggressive Periodontitis and Ectrodactyly-Ectodermal Dysplasia-Cleft Syndrome
Case Report Association of generalized aggressive periodontitis and ectrodactyly-ectodermal dysplasia-cleft syndrome Rosamma Joseph, Sameera G. Nath Department of Periodontics, Government Dental College, Medical College P.O., Calicut, Kerala, India p63 , the predominant isotope in epithelial basal cell Ectrodactyly-ectodermal dysplasia-cleft (EEC) syndrome is an autosomal dominant disorder characterized by the triad layers, is responsible for major anomalies found in of ectrodactyly, ectodermal dysplasia, and facial clefting. patients with EEC syndrome.[3] Even though literature has documented the association of various genetic disorders with aggressive periodontitis, the Several case reports[4-6] of EEC syndrome has been periodontal manifestations in patients with EEC syndrome published pertaining to the dermatological, urogenital, have never been addressed. This case report presents the periodontal status of three patients in a family with and dental/oral manifestations of EEC syndrome. EEC syndrome. The presence of generalized aggressive Oral manifestations of patients with EEC syndrome periodontitis was noticed in these patients. EEC syndrome reported so far include cleft lip or palate, cleft palate could be a new addition to the group of genetic disorders associated with aggressive periodontitis. alone, hypodontia, microdontia, anodontia, xerostomia contributing to high caries rate with dry granulomatous Key words: Aggressive periodontitis, case lesions on lips, and parotid duct atresia. The periodontal report,ectrodactyly-ectodermal dysplasia-cleft syndrome manifestations in EEC syndrome has not been documented in literature. Introduction Although, literature[7,8] has documented the association of various genetic disorders with aggressive periodontitis, the periodontal manifestations in patients with EEC Ectrodactyly-ectodermal dysplasia-cleft syndrome syndrome have never been addressed. -

EUROCAT Syndrome Guide
JRC - Central Registry european surveillance of congenital anomalies EUROCAT Syndrome Guide Definition and Coding of Syndromes Version July 2017 Revised in 2016 by Ingeborg Barisic, approved by the Coding & Classification Committee in 2017: Ester Garne, Diana Wellesley, David Tucker, Jorieke Bergman and Ingeborg Barisic Revised 2008 by Ingeborg Barisic, Helen Dolk and Ester Garne and discussed and approved by the Coding & Classification Committee 2008: Elisa Calzolari, Diana Wellesley, David Tucker, Ingeborg Barisic, Ester Garne The list of syndromes contained in the previous EUROCAT “Guide to the Coding of Eponyms and Syndromes” (Josephine Weatherall, 1979) was revised by Ingeborg Barisic, Helen Dolk, Ester Garne, Claude Stoll and Diana Wellesley at a meeting in London in November 2003. Approved by the members EUROCAT Coding & Classification Committee 2004: Ingeborg Barisic, Elisa Calzolari, Ester Garne, Annukka Ritvanen, Claude Stoll, Diana Wellesley 1 TABLE OF CONTENTS Introduction and Definitions 6 Coding Notes and Explanation of Guide 10 List of conditions to be coded in the syndrome field 13 List of conditions which should not be coded as syndromes 14 Syndromes – monogenic or unknown etiology Aarskog syndrome 18 Acrocephalopolysyndactyly (all types) 19 Alagille syndrome 20 Alport syndrome 21 Angelman syndrome 22 Aniridia-Wilms tumor syndrome, WAGR 23 Apert syndrome 24 Bardet-Biedl syndrome 25 Beckwith-Wiedemann syndrome (EMG syndrome) 26 Blepharophimosis-ptosis syndrome 28 Branchiootorenal syndrome (Melnick-Fraser syndrome) 29 CHARGE -

Emanuel Syndrome Information
CHROMOSOME DISORDER OUTREACH EMANUEL SYNDROME INFORMATION Emanuel Syndrome All chromosomal syndromes are caused by deletions (duplications, trisomies) of one chromosome. The only exception is Emanuel syndrome (ES), caused by the cumulative action of partial trisomies for two chromosomes: the distal segment of chromosome 11q and the proximal segment of chromosome 22q. Most non-Robertsonian translocations are unique. Translocation t(11;22) is an exception. It is the most common non-Robertsonian translocation in humans. The structural similarity of the 22q11.2 segment and the 11q23 segment facilitates the exchange between these areas with formation of t(11;22)(q23; q11.2). As a result of the segregation 3:1 which is almost constant in this translocation1 the patient will have an additional chromosome der(22) which includes part of chromosome 22 (short arm, centromere and proximal segment of 11q [up to 22q11.2]) and the distal part of chromosome 11 (from 11q23 to the end of the long arm). This syndrome is usually called Emanuel syndrome after Beverly Emanuel who studied this condition for many years2. There is no doubt that several patients described in the early cytogenetic literature as persons with “trisomy 22” actually had ES. Sometimes the term “supernumerary der(22)t(11;22)” syndrome is used to describe this disorder3. page 1 ES is a relatively common pathology. At least 400 patients have been reported so far. The clinical manifestations of the syndrome include severe developmental delay, defects of the face, heart, kidneys and other systems. Most patients have prenatal hypoplasia and their physical parameters (weight, height) are much below those of their healthy peers.