Accelerating Matchmaking of Novel Dysmorphology Syndromes Through Clinical and Genomic Characterization of a Large Cohort
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PARSANA-DISSERTATION-2020.Pdf
DECIPHERING TRANSCRIPTIONAL PATTERNS OF GENE REGULATION: A COMPUTATIONAL APPROACH by Princy Parsana A dissertation submitted to The Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July, 2020 © 2020 Princy Parsana All rights reserved Abstract With rapid advancements in sequencing technology, we now have the ability to sequence the entire human genome, and to quantify expression of tens of thousands of genes from hundreds of individuals. This provides an extraordinary opportunity to learn phenotype relevant genomic patterns that can improve our understanding of molecular and cellular processes underlying a trait. The high dimensional nature of genomic data presents a range of computational and statistical challenges. This dissertation presents a compilation of projects that were driven by the motivation to efficiently capture gene regulatory patterns in the human transcriptome, while addressing statistical and computational challenges that accompany this data. We attempt to address two major difficulties in this domain: a) artifacts and noise in transcriptomic data, andb) limited statistical power. First, we present our work on investigating the effect of artifactual variation in gene expression data and its impact on trans-eQTL discovery. Here we performed an in-depth analysis of diverse pre-recorded covariates and latent confounders to understand their contribution to heterogeneity in gene expression measurements. Next, we discovered 673 trans-eQTLs across 16 human tissues using v6 data from the Genotype Tissue Expression (GTEx) project. Finally, we characterized two trait-associated trans-eQTLs; one in Skeletal Muscle and another in Thyroid. Second, we present a principal component based residualization method to correct gene expression measurements prior to reconstruction of co-expression networks. -

DNA Breakpoint Assay Reveals a Majority of Gross Duplications Occur in Tandem Reducing VUS Classifications in Breast Cancer Predisposition Genes
© American College of Medical Genetics and Genomics ARTICLE Corrected: Correction DNA breakpoint assay reveals a majority of gross duplications occur in tandem reducing VUS classifications in breast cancer predisposition genes Marcy E. Richardson, PhD1, Hansook Chong, PhD1, Wenbo Mu, MS1, Blair R. Conner, MS1, Vickie Hsuan, MS1, Sara Willett, MS1, Stephanie Lam, MS1, Pei Tsai, CGMBS, MB (ASCP)1, Tina Pesaran, MS, CGC1, Adam C. Chamberlin, PhD1, Min-Sun Park, PhD1, Phillip Gray, PhD1, Rachid Karam, MD, PhD1 and Aaron Elliott, PhD1 Purpose: Gross duplications are ambiguous in terms of clinical cohort, while the remainder have unknown tandem status. Among interpretation due to the limitations of the detection methods that the tandem gross duplications that were eligible for reclassification, cannot infer their context, namely, whether they occur in tandem or 95% of them were upgraded to pathogenic. are duplicated and inserted elsewhere in the genome. We Conclusion: DBA is a novel, high-throughput, NGS-based method investigated the proportion of gross duplications occurring in that informs the tandem status, and thereby the classification of, tandem in breast cancer predisposition genes with the intent of gross duplications. This method revealed that most gross duplica- informing their classifications. tions in the investigated genes occurred in tandem and resulted in a Methods: The DNA breakpoint assay (DBA) is a custom, paired- pathogenic classification, which helps to secure the necessary end, next-generation sequencing (NGS) method designed to treatment options for their carriers. capture and detect deep-intronic DNA breakpoints in gross duplications in BRCA1, BRCA2, ATM, CDH1, PALB2, and CHEK2. Genetics in Medicine (2019) 21:683–693; https://doi.org/10.1038/s41436- Results: DBA allowed us to ascertain breakpoints for 44 unique 018-0092-7 gross duplications from 147 probands. -

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

Crouzon Syndrome with Ophthalmological Complications
Journal of Rawalpindi Medical College (JRMC); 2012;16(1):80-81 Case Report Crouzon Syndrome With Ophthalmological Complications Rahela Nasir Paediatric Department, Capital Hospital, CDA Islamabad. Crouzon Syndrome is characterized by premature bilateral optic atrophy. Other facial features included a craniosynostosis. It has an autosomal dominant inheritance prominent nose and deep and narrow palate. No but represents fresh mutation also. Other craniofacial digital abnormalities were seen. No dental aplasia was abnormalities include ocular proptosis caused by shallow present. Systemic examination revealed no orbits with or without divergent strabismus. There may be abnormality. increased intracranial pressure for which surgical morcellation procedures are indicated. A case of He was diagnosed as a case of Crouzon syndrome craniosynostosis is reported which is diagnosed as Crouzon with ocular complications on clinical basis. He was Syndrome with ocular complications on clinical grounds. investigated for microcephaly and suspected Split craniotomy was performed by a neurosurgeon to craniosynostosis. Radiographs of skull showed small relieve raised intracranial pressure and to enhance brain sized skull with early closure of sutures and fontanelle growth. Crouzon Syndrome was originally described in 1912 suggestive of craniosynstosis(Fig 2). A hammered- by Crouzon in a mother and her daughter. It is an autosomal silver (beaten metal/ copper beaten) appearance was dominant inherited disorder but represents fresh mutation also seen due to raised intracranial pressure and also. Crouzon syndrome is characterized by premature compression of the developing brain on the fused craniosynostosis which is quite variable but the coronal suture is nearly always bilaterally involved. Craniofacial bone. CT scan brain with contrast was done which abnormalities include brachycephaly, shallow orbits and confirmed the features suggestive of craniosynostosis maxillary hypoplasia. -
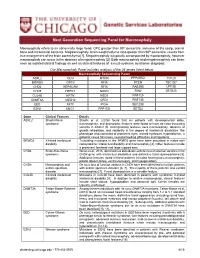
Macrocephaly Information Sheet 6-13-19
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Craniosynostosis and Related Disorders Mark S
CLINICAL REPORT Guidance for the Clinician in Rendering Pediatric Care Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S. Dias, MD, FAAP, FAANS,a Thomas Samson, MD, FAAP,b Elias B. Rizk, MD, FAAP, FAANS,a Lance S. Governale, MD, FAAP, FAANS,c Joan T. Richtsmeier, PhD,d SECTION ON NEUROLOGIC SURGERY, SECTION ON PLASTIC AND RECONSTRUCTIVE SURGERY Pediatric care providers, pediatricians, pediatric subspecialty physicians, and abstract other health care providers should be able to recognize children with abnormal head shapes that occur as a result of both synostotic and aSection of Pediatric Neurosurgery, Department of Neurosurgery and deformational processes. The purpose of this clinical report is to review the bDivision of Plastic Surgery, Department of Surgery, College of characteristic head shape changes, as well as secondary craniofacial Medicine and dDepartment of Anthropology, College of the Liberal Arts characteristics, that occur in the setting of the various primary and Huck Institutes of the Life Sciences, Pennsylvania State University, State College, Pennsylvania; and cLillian S. Wells Department of craniosynostoses and deformations. As an introduction, the physiology and Neurosurgery, College of Medicine, University of Florida, Gainesville, genetics of skull growth as well as the pathophysiology underlying Florida craniosynostosis are reviewed. This is followed by a description of each type of Clinical reports from the American Academy of Pediatrics benefit from primary craniosynostosis (metopic, unicoronal, bicoronal, sagittal, lambdoid, expertise and resources of liaisons and internal (AAP) and external reviewers. However, clinical reports from the American Academy of and frontosphenoidal) and their resultant head shape changes, with an Pediatrics may not reflect the views of the liaisons or the emphasis on differentiating conditions that require surgical correction from organizations or government agencies that they represent. -

Blueprint Genetics Craniosynostosis Panel
Craniosynostosis Panel Test code: MA2901 Is a 38 gene panel that includes assessment of non-coding variants. Is ideal for patients with craniosynostosis. About Craniosynostosis Craniosynostosis is defined as the premature fusion of one or more cranial sutures leading to secondary distortion of skull shape. It may result from a primary defect of ossification (primary craniosynostosis) or, more commonly, from a failure of brain growth (secondary craniosynostosis). Premature closure of the sutures (fibrous joints) causes the pressure inside of the head to increase and the skull or facial bones to change from a normal, symmetrical appearance resulting in skull deformities with a variable presentation. Craniosynostosis may occur in an isolated setting or as part of a syndrome with a variety of inheritance patterns and reccurrence risks. Craniosynostosis occurs in 1/2,200 live births. Availability 4 weeks Gene Set Description Genes in the Craniosynostosis Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ALPL Odontohypophosphatasia, Hypophosphatasia perinatal lethal, AD/AR 78 291 infantile, juvenile and adult forms ALX3 Frontonasal dysplasia type 1 AR 8 8 ALX4 Frontonasal dysplasia type 2, Parietal foramina AD/AR 15 24 BMP4 Microphthalmia, syndromic, Orofacial cleft AD 8 39 CDC45 Meier-Gorlin syndrome 7 AR 10 19 EDNRB Hirschsprung disease, ABCD syndrome, Waardenburg syndrome AD/AR 12 66 EFNB1 Craniofrontonasal dysplasia XL 28 116 ERF Craniosynostosis 4 AD 17 16 ESCO2 SC phocomelia syndrome, Roberts syndrome -

REVIEW ARTICLE the Genetics of Autism
REVIEW ARTICLE The Genetics of Autism Rebecca Muhle, BA*; Stephanie V. Trentacoste, BA*; and Isabelle Rapin, MD‡ ABSTRACT. Autism is a complex, behaviorally de- tribution of a few well characterized X-linked disorders, fined, static disorder of the immature brain that is of male-to-male transmission in a number of families rules great concern to the practicing pediatrician because of an out X-linkage as the prevailing mode of inheritance. The astonishing 556% reported increase in pediatric preva- recurrence rate in siblings of affected children is ϳ2% to lence between 1991 and 1997, to a prevalence higher than 8%, much higher than the prevalence rate in the general that of spina bifida, cancer, or Down syndrome. This population but much lower than in single-gene diseases. jump is probably attributable to heightened awareness Twin studies reported 60% concordance for classic au- and changing diagnostic criteria rather than to new en- tism in monozygotic (MZ) twins versus 0 in dizygotic vironmental influences. Autism is not a disease but a (DZ) twins, the higher MZ concordance attesting to ge- syndrome with multiple nongenetic and genetic causes. netic inheritance as the predominant causative agent. By autism (the autistic spectrum disorders [ASDs]), we Reevaluation for a broader autistic phenotype that in- mean the wide spectrum of developmental disorders cluded communication and social disorders increased characterized by impairments in 3 behavioral domains: 1) concordance remarkably from 60% to 92% in MZ twins social interaction; 2) language, communication, and and from 0% to 10% in DZ pairs. This suggests that imaginative play; and 3) range of interests and activities. -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

Macrophage Activation JUNB Is a Key Transcriptional Modulator Of
JUNB Is a Key Transcriptional Modulator of Macrophage Activation Mary F. Fontana, Alyssa Baccarella, Nidhi Pancholi, Miles A. Pufall, De'Broski R. Herbert and Charles C. Kim This information is current as of October 2, 2021. J Immunol 2015; 194:177-186; Prepublished online 3 December 2014; doi: 10.4049/jimmunol.1401595 http://www.jimmunol.org/content/194/1/177 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2014/12/03/jimmunol.140159 Material 5.DCSupplemental References This article cites 40 articles, 7 of which you can access for free at: http://www.jimmunol.org/content/194/1/177.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 2, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2014 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology JUNB Is a Key Transcriptional Modulator of Macrophage Activation Mary F. Fontana,* Alyssa Baccarella,* Nidhi Pancholi,* Miles A. -

Sotos Syndrome
European Journal of Human Genetics (2007) 15, 264–271 & 2007 Nature Publishing Group All rights reserved 1018-4813/07 $30.00 www.nature.com/ejhg PRACTICAL GENETICS In association with Sotos syndrome Sotos syndrome is an autosomal dominant condition characterised by a distinctive facial appearance, learning disability and overgrowth resulting in tall stature and macrocephaly. In 2002, Sotos syndrome was shown to be caused by mutations and deletions of NSD1, which encodes a histone methyltransferase implicated in chromatin regulation. More recently, the NSD1 mutational spectrum has been defined, the phenotype of Sotos syndrome clarified and diagnostic and management guidelines developed. Introduction In brief Sotos syndrome was first described in 1964 by Juan Sotos Sotos syndrome is characterised by a distinctive facial and the major diagnostic criteria of a distinctive facial appearance, learning disability and childhood over- appearance, childhood overgrowth and learning disability growth. were established in 1994 by Cole and Hughes.1,2 In 2002, Sotos syndrome is associated with cardiac anomalies, cloning of the breakpoints of a de novo t(5;8)(q35;q24.1) renal anomalies, seizures and/or scoliosis in B25% of translocation in a child with Sotos syndrome led to the cases and a broad variety of additional features occur discovery that Sotos syndrome is caused by haploinsuffi- less frequently. ciency of the Nuclear receptor Set Domain containing NSD1 abnormalities, such as truncating mutations, protein 1 gene, NSD1.3 Subsequently, extensive analyses of missense mutations in functional domains, partial overgrowth cases have shown that intragenic NSD1 muta- gene deletions and 5q35 microdeletions encompass- tions and 5q35 microdeletions encompassing NSD1 cause ing NSD1, are identifiable in the majority (490%) of 490% of Sotos syndrome cases.4–10 In addition, NSD1 Sotos syndrome cases. -

KBG Syndrome Presenting with Brachydactyly Type E
Bone 123 (2019) 18–22 Contents lists available at ScienceDirect Bone journal homepage: www.elsevier.com/locate/bone Case Report KBG syndrome presenting with brachydactyly type E T ⁎ Renata Libiantoa,b,c, , Kathy HC Wud,e,f,g, Sophie Deveryd, John A Eismana,b,f,h, Jackie R Centera,b,f,h a Bone Division, Garvan Institute of Medical Research, Sydney, Australia b Department of Endocrinology, St Vincent's Hospital Sydney, Australia c Department of Medicine, The University of Melbourne, Australia d Clinical Genomics Unit, St Vincent's Hospital Sydney, Australia e Discipline of Genetic Medicine, University of Sydney, Australia f School of Medicine, UNSW, Sydney, Australia g Genomics and Epigenetics Division, Garvan Institute of Medical Research, Sydney, Australia h School of Medicine Sydney, University of Notre Dame, Australia ARTICLE INFO ABSTRACT Keywords: We report the case of a young woman who presented at age 10 years with height on the tenth centile, bra- KBG syndrome chydactyly type E and mild developmental delay. Biochemistry and hormonal profiles were normal. Differential ANKRD11 gene diagnoses considered included Albright hereditary osteodystrophy without hormone resistance (a.k.a pseu- Skeletal disorder dopseudohypoparathyroidism), 2q37 microdeletion syndrome and acrodysostosis. She had a normal karyotype Brachydactyly type E and normal FISH of 2q37. Whole genome sequencing (WGS) identified a mutation in the ANKRD11 gene as- sociated with KBG syndrome. We review the clinical features of the genetic syndromes considered, and suggest KBG syndrome be considered in patients presenting with syndromic brachydactyly type E, especially if short stature and developmental delay are also present. 1. Case support in a mainstream school for specific learning difficulties.