Cracking the Odor Code: Molecular and Cellular Deconstruction of the Olfactory Circuit of Drosophila Larvae Kenta Asahina
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Medical Review Officer Manual
Department of Health and Human Services Substance Abuse and Mental Health Services Administration Center for Substance Abuse Prevention Medical Review Officer Manual for Federal Agency Workplace Drug Testing Programs EFFECTIVE OCTOBER 1, 2010 Note: This manual applies to Federal agency drug testing programs that come under Executive Order 12564 dated September 15, 1986, section 503 of Public Law 100-71, 5 U.S.C. section 7301 note dated July 11, 1987, and the Department of Health and Human Services Mandatory Guidelines for Federal Workplace Drug Testing Programs (73 FR 71858) dated November 25, 2008 (effective October 1, 2010). This manual does not apply to specimens submitted for testing under U.S. Department of Transportation (DOT) Procedures for Transportation Workplace Drug and Alcohol Testing Programs (49 CFR Part 40). The current version of this manual and other information including MRO Case Studies are available on the Drug Testing page under Medical Review Officer (MRO) Resources on the SAMHSA website: http://www.workplace.samhsa.gov Previous Versions of this Manual are Obsolete 3 Table of Contents Chapter 1. The Medical Review Officer (MRO)........................................................................... 6 Chapter 2. The Federal Drug Testing Custody and Control Form ................................................ 7 Chapter 3. Urine Drug Testing ...................................................................................................... 9 A. Federal Workplace Drug Testing Overview.................................................................. -

Chemoreception
Senses 5 SENSES live version • discussion • edit lesson • comment • report an error enses are the physiological methods of perception. The senses and their operation, classification, Sand theory are overlapping topics studied by a variety of fields. Sense is a faculty by which outside stimuli are perceived. We experience reality through our senses. A sense is a faculty by which outside stimuli are perceived. Many neurologists disagree about how many senses there actually are due to a broad interpretation of the definition of a sense. Our senses are split into two different groups. Our Exteroceptors detect stimulation from the outsides of our body. For example smell,taste,and equilibrium. The Interoceptors receive stimulation from the inside of our bodies. For instance, blood pressure dropping, changes in the gluclose and Ph levels. Children are generally taught that there are five senses (sight, hearing, touch, smell, taste). However, it is generally agreed that there are at least seven different senses in humans, and a minimum of two more observed in other organisms. Sense can also differ from one person to the next. Take taste for an example, what may taste great to me will taste awful to someone else. This all has to do with how our brains interpret the stimuli that is given. Chemoreception The senses of Gustation (taste) and Olfaction (smell) fall under the category of Chemoreception. Specialized cells act as receptors for certain chemical compounds. As these compounds react with the receptors, an impulse is sent to the brain and is registered as a certain taste or smell. Gustation and Olfaction are chemical senses because the receptors they contain are sensitive to the molecules in the food we eat, along with the air we breath. -

Understanding Sensory Processing: Looking at Children's Behavior Through the Lens of Sensory Processing
Understanding Sensory Processing: Looking at Children’s Behavior Through the Lens of Sensory Processing Communities of Practice in Autism September 24, 2009 Charlottesville, VA Dianne Koontz Lowman, Ed.D. Early Childhood Coordinator Region 5 T/TAC James Madison University MSC 9002 Harrisonburg, VA 22807 [email protected] ______________________________________________________________________________ Dianne Koontz Lowman/[email protected]/2008 Page 1 Looking at Children’s Behavior Through the Lens of Sensory Processing Do you know a child like this? Travis is constantly moving, pushing, or chewing on things. The collar of his shirt and coat are always wet from chewing. When talking to people, he tends to push up against you. Or do you know another child? Sierra does not like to be hugged or kissed by anyone. She gets upset with other children bump up against her. She doesn’t like socks with a heel or toe seam or any tags on clothes. Why is Travis always chewing? Why doesn’t Sierra liked to be touched? Why do children react differently to things around them? These children have different ways of reacting to the things around them, to sensations. Over the years, different terms (such as sensory integration) have been used to describe how children deal with the information they receive through their senses. Currently, the term being used to describe children who have difficulty dealing with input from their senses is sensory processing disorder. _____________________________________________________________________ Sensory Processing Disorder -

The Olfactory Bulb Theta Rhythm Follows All Frequencies of Diaphragmatic Respiration in the Freely Behaving Rat
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Frontiers - Publisher Connector ORIGINAL RESEARCH ARTICLE published: 11 June 2014 BEHAVIORAL NEUROSCIENCE doi: 10.3389/fnbeh.2014.00214 The olfactory bulb theta rhythm follows all frequencies of diaphragmatic respiration in the freely behaving rat Daniel Rojas-Líbano 1,2†, Donald E. Frederick 2,3, José I. Egaña 4 and Leslie M. Kay 1,2,3* 1 Committee on Neurobiology, The University of Chicago, Chicago, IL, USA 2 Institute for Mind and Biology, The University of Chicago, Chicago, IL, USA 3 Department of Psychology, The University of Chicago, Chicago, IL, USA 4 Departamento de Anestesiología y Reanimación, Facultad de Medicina, Universidad de Chile, Santiago, Chile Edited by: Sensory-motor relationships are part of the normal operation of sensory systems. Sensing Donald A. Wilson, New York occurs in the context of active sensor movement, which in turn influences sensory University School of Medicine, USA processing. We address such a process in the rat olfactory system. Through recordings of Reviewed by: the diaphragm electromyogram (EMG), we monitored the motor output of the respiratory Thomas A. Cleland, Cornell University, USA circuit involved in sniffing behavior, simultaneously with the local field potential (LFP) of Emmanuelle Courtiol, New York the olfactory bulb (OB) in rats moving freely in a familiar environment, where they display University Langone Medical Center, a wide range of respiratory frequencies. We show that the OB LFP represents the sniff USA cycle with high reliability at every sniff frequency and can therefore be used to study the *Correspondence: neural representation of motor drive in a sensory cortex. -

Color, Taste, and Odor: What You Should Know
Color, Taste, and Odor: What you should know From time to time the MassDEP receives consumer questions or complaints regarding the look, taste or the odor of drinking water. Listed below are common problems with drinking water and their most common causes. Please note that a particular problem in your drinking water may be the result of a cause not listed here; the only way to confirm a cause is to have a certified lab analyze the water and discuss the results with drinking water professional. If you receive water from a public drinking water system it is important to contact the Public Water Supply (PWS) before having a laboratory analyze the water. Information on private water testing is available. Filtering or treating the water may remedy persistent problems; however MassDEP does not recommend filtering or treating your water supply if your water is supplied by a MassDEP- approved PWS. MassDEP also does not regulate or recommend specific treatment systems for private home use. If you decide to use a filtration or treatment device in your home, the Department strongly encourages you to contact National Sanitation Foundation (NSF) for a list of approved devices. If you purchase a treatment device for private home use MassDEP also strongly recommends that it is maintained and provide active maintenance according to the manufacturer's instructions. Failure to maintain the equipment properly may make treatment ineffective and/or may create the potential for contamination. Common problems with drinking water are grouped into three categories: Color problems Taste / odor problems Particles in water If the problem with your water is not described here, if you are on a public water system please contact the public water department in your city or town or the MassDEP Drinking Water Program at your nearest regional MassDEP office. -

Special Issue “Olfaction: from Genes to Behavior”
G C A T T A C G G C A T genes Editorial Special Issue “Olfaction: From Genes to Behavior” Edgar Soria-Gómez 1,2,3 1 Department of Neurosciences, University of the Basque Country UPV/EHU, 48940 Leioa, Spain; [email protected] or [email protected] 2 Achucarro Basque Center for Neuroscience, Science Park of the UPV/EHU, 48940 Leioa, Spain 3 IKERBASQUE, Basque Foundation for Science, 48013 Bilbao, Spain Received: 12 June 2020; Accepted: 15 June 2020; Published: 15 June 2020 The senses dictate how the brain represents the environment, and this representation is the basis of how we act in the world. Among the five senses, olfaction is maybe the most mysterious and underestimated one, probably because a large part of the olfactory information is processed at the unconscious level in humans [1–4]. However, it is undeniable the influence of olfaction in the control of behavior and cognitive processes. Indeed, many studies demonstrate a tight relationship between olfactory perception and behavior [5]. For example, olfactory cues are determinant for partner selection [6,7], parental care [8,9], and feeding behavior [10–13], and the sense of smell can even contribute to emotional responses, cognition and mood regulation [14,15]. Accordingly, it has been shown that a malfunctioning of the olfactory system could be causally associated with the occurrence of important diseases, such as neuropsychiatric depression or feeding-related disorders [16,17]. Thus, a clear identification of the biological mechanisms involved in olfaction is key in the understanding of animal behavior in physiological and pathological conditions. -
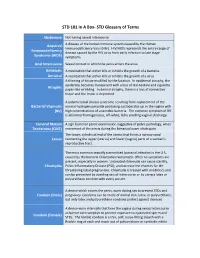
STD Glossary of Terms
STD 101 In A Box- STD Glossary of Terms Abstinence Not having sexual intercourse Acquired A disease of the human immune system caused by the Human Immunodeficiency Virus (HIV). HIV/AIDS represents the entire range of Immunodeficiency disease caused by the HIV virus from early infection to late stage Syndrome (AIDS) symptoms. Anal Intercourse Sexual contact in which the penis enters the anus. Antibiotic A medication that either kills or inhibits the growth of a bacteria. Antiviral A medication that either kills or inhibits the growth of a virus. A thinning of tissue modified by the location. In epidermal atrophy, the epidermis becomes transparent with a loss of skin texture and cigarette Atrophic paper-like wrinkling. In dermal atrophy, there is a loss of connective tissue and the lesion is depressed. A polymicrobial clinical syndrome resulting from replacement of the Bacterial Vaginosis normal hydrogen peroxide producing Lactobacillus sp. in the vagina with (BV) high concentrations of anaerobic bacteria. The common symptom of BV is abnormal homogeneous, off-white, fishy smelling vaginal discharge. Cervical Motion A sign found on pelvic examination suggestive of pelvic pathology; when Tenderness (CMT) movement of the cervix during the bimanual exam elicits pain. The lower, cylindrical end of the uterus that forms a narrow canal Cervix connecting the upper (uterus) and lower (vagina) parts of a woman's reproductive tract. The most common sexually transmitted bacterial infection in the U.S., caused by the bacteria Chlamydia trachomatis. Often no symptoms are present, especially in women. Untreated chlamydia can cause sterility, Chlamydia Pelvic Inflammatory Disease (PID), and increase the chances for life- threatening tubal pregnancies. -

A Pheromone Antagonist Liberates Female Sea Lamprey from a Sensory Trap to Enable Reliable Communication
A pheromone antagonist liberates female sea lamprey from a sensory trap to enable reliable communication Tyler J. Buchingera,1, Anne M. Scotta,1, Skye D. Fissettea, Cory O. Branta,2, Mar Huertasa,3,KeLia,4, Nicholas S. Johnsonb, and Weiming Lia,5 aDepartment of Fisheries and Wildlife, Michigan State University, East Lansing, MI 48824; and bHammond Bay Biological Station, US Geological Survey, Millersburg, MI 49759 Edited by John G. Hildebrand, University of Arizona, Tucson, AZ, and approved February 21, 2020 (received for review December 12, 2019) The evolution of male signals and female preferences remains a subsequent preference evolution precludes a full understanding central question in the study of animal communication. The sensory of how receiver biases shape communication. trap model suggests males evolve signals that mimic cues used Sea lamprey (Petromyzon marinus) are a useful model to track in nonsexual contexts and thus manipulate female behavior to the evolutionary trajectory of communication that originated via generate mating opportunities. Much evidence supports the sen- a sensory trap. Chemical cues and pheromones guide sea lam- sory trap model, but how females glean reliable information from prey behavior throughout their complex life history, especially both mimetic signals and their model cues remains unknown. We during the terminal reproductive phase (21). After parasitizing discovered a mechanism whereby a manipulative male signal guides fish in lakes or the Atlantic Ocean, prespawning sea lamprey reliable communication in sea lamprey (Petromyzon marinus). Mi- migrate into streams following chemical cues released by larvae gratory sea lamprey follow a larval cue into spawning streams; once residing in nursery habitats near spawning grounds. -

Chapter 17: the Special Senses
Chapter 17: The Special Senses I. An Introduction to the Special Senses, p. 550 • The state of our nervous systems determines what we perceive. 1. For example, during sympathetic activation, we experience a heightened awareness of sensory information and hear sounds that would normally escape our notice. 2. Yet, when concentrating on a difficult problem, we may remain unaware of relatively loud noises. • The five special senses are: olfaction, gustation, vision, equilibrium, and hearing. II. Olfaction, p. 550 Objectives 1. Describe the sensory organs of smell and trace the olfactory pathways to their destinations in the brain. 2. Explain what is meant by olfactory discrimination and briefly describe the physiology involved. • The olfactory organs are located in the nasal cavity on either side of the nasal septum. Figure 17-1a • The olfactory organs are made up of two layers: the olfactory epithelium and the lamina propria. • The olfactory epithelium contains the olfactory receptors, supporting cells, and basal (stem) cells. Figure 17–1b • The lamina propria consists of areolar tissue, numerous blood vessels, nerves, and olfactory glands. • The surfaces of the olfactory organs are coated with the secretions of the olfactory glands. Olfactory Receptors, p. 551 • The olfactory receptors are highly modified neurons. • Olfactory reception involves detecting dissolved chemicals as they interact with odorant-binding proteins. Olfactory Pathways, p. 551 • Axons leaving the olfactory epithelium collect into 20 or more bundles that penetrate the cribriform plate of the ethmoid bone to reach the olfactory bulbs of the cerebrum where the first synapse occurs. • Axons leaving the olfactory bulb travel along the olfactory tract to reach the olfactory cortex, the hypothalamus, and portions of the limbic system. -

Taste and Odor Control, but Use As a Control Chemical Must Be Evaluated Carefully Due to the Formation of Thms and Chlorophenol When Organics Are Present
Taste and Odor Most customers judge the quality of drinking water by taste and odor. If the customer is satisfied with these qualities, it is assumed the water is safe to drink. Many harmful contaminants in water cannot be detected due to taste or smell and many of the contaminants found in drinking water that have a detectable taste or odor are not harmful. Sources of taste and odor problems can be found in surface water and groundwater. Source water protection involves the prevention of contaminants from entering the source. Surface water or groundwater may become contaminated by pollutants such as gasoline, industrial solvents or a wide variety of volatile organics. The removal of contaminants from surface water or groundwater is costly and may involve the use of aeration, powdered activated carbon, or both. If taste and odor must be controlled at the treatment plant, oxidation, aeration and adsorption can be effective in reducing taste and odor, and improved coagulation filtration. ODOR MEASUREMENTS One of the most common methods for measuring odor in water is the threshold odor test. It involves a series of flasks presented to an observer, who is told that some of the samples contain odors and that the series is arranged in order of increasing concentrations. The observer is also given a known odor-free blank for reference during the test. The observer compares the flasks in ascending order with the blank and then notes whether an odor is detected in any sample flask. Individuals vary in their reactions to certain types of odors. An odor stimulus that is agreeable to one may be disagreeable to another. -

Feedforward and Feedback Signals in the Olfactory System Srimoy Chakraborty University of Arkansas, Fayetteville
University of Arkansas, Fayetteville ScholarWorks@UARK Theses and Dissertations 5-2019 Feedforward and Feedback Signals in the Olfactory System Srimoy Chakraborty University of Arkansas, Fayetteville Follow this and additional works at: https://scholarworks.uark.edu/etd Part of the Behavioral Neurobiology Commons, Bioelectrical and Neuroengineering Commons, Cognitive Neuroscience Commons, Engineering Physics Commons, Health and Medical Physics Commons, and the Systems Neuroscience Commons Recommended Citation Chakraborty, Srimoy, "Feedforward and Feedback Signals in the Olfactory System" (2019). Theses and Dissertations. 3145. https://scholarworks.uark.edu/etd/3145 This Dissertation is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of ScholarWorks@UARK. For more information, please contact [email protected]. Feedforward and Feedback Signals in the Olfactory System A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Physics by Srimoy Chakraborty University of Calcutta Bachelor of Science in Physics, 2008 University of Arkansas Master of Science in Physics, 2015 May 2019 University of Arkansas This dissertation is approved for recommendation to the Graduate Council. _________________________________ Woodrow Shew, Ph.D. Dissertation Director _________________________________ Wayne J. Kuenzel, Ph.D. Committee Member _________________________________ Jiali Li, Ph.D. Committee Member _________________________________ Pradeep Kumar, Ph.D. Committee Member Abstract The conglomeration of myriad activities in neural systems often results in prominent oscillations . The primary goal of the research presented in this thesis was to study effects of sensory stimulus on the olfactory system of rats, focusing on the olfactory bulb (OB) and the anterior piriform cortex (aPC). -

The Role of Chemoreceptor Evolution in Behavioral Change Cande, Prud’Homme and Gompel 153
Available online at www.sciencedirect.com Smells like evolution: the role of chemoreceptor evolution in behavioral change Jessica Cande, Benjamin Prud’homme and Nicolas Gompel In contrast to physiology and morphology, our understanding success. How an organism interacts with its environment of how behaviors evolve is limited.This is a challenging task, as can be divided into three parts: first, the sensory percep- it involves the identification of both the underlying genetic tion of diverse auditory, visual, tactile, chemosensory or basis and the resultant physiological changes that lead to other cues; second, the processing of this information by behavioral divergence. In this review, we focus on the central nervous system (CNS), leading to a repres- chemosensory systems, mostly in Drosophila, as they are one entation of the sensory signal; and third, a behavioral of the best-characterized components of the nervous system response. Thus, behaviors could evolve either through in model organisms, and evolve rapidly between species. We changes in the peripheral nervous system (PNS) (e.g. examine the hypothesis that changes at the level of [1 ]), or through changes in higher-order neural circuitry chemosensory systems contribute to the diversification of (Figure 1). While the latter remain elusive, recent work behaviors. In particular, we review recent progress in on chemosensation in insects illustrates how the PNS understanding how genetic changes between species affect shapes behavioral evolution. chemosensory systems and translate into divergent behaviors. A major evolutionary trend is the rapid Chemosensation in insects depends on three classes of diversification of the chemoreceptor repertoire among receptors expressed in peripheral neurons housed in species.