A Molecular Phylogenetic Study of Graptopetalum (Crassulaceae) Based on Ets, Its, Rpl16, and Trnl-F Nucleotide Sequences1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Succulent Manual- a Guide to Care and Repair for All Climates
Contents Title page Dedication Preface Chapter 1: Basic Tips Color and Form Light Watering Soil and Fertilizer Containers Growth, Dormancy Flowers, Seeds Chapter 2: Make More Sucs Intro Propagation by Leaf By Division By Cuttings By Seed Chapter 3: Succulent SOS Symptoms Signs in Leaves Signs in Stems Signs in Roots Signs of Fungal Diseases Signs of Common Pests Take Action Increase Hydration Reduce Hydration Increase Light Decrease Light Color Repair Etiolation Repair Stem Rot Root Rot Fungi Pests Mealybugs Aphids Spider Mites Scale Slugs and Snails Caterpillars Birds and Mammals Physical Damage Chapter 4: Regional Tips Intro Lack of Sunlight Hot-Humid Hot-Arid Cold-Humid, Semi-arid to Arid Mixed Indoors Chapter 5: Identification How to ID your plants Chapter 6: Genus Tips Aloe Cacti Echeveria Haworthia Euphorbia Lithops Mesembs Other Popular Varieties Chapter 7: In-ground Succulents Care and Garden Construction Chapter 8: Tasks and Projects Potting up plants Projects Cleaning your plants Drilling for drainage DIY shelves Logging and Journaling Leaving town Chapter 9: Buying Guide Plants Supplies Knowledge Bank Succulent Anatomy Glossary About the Author The Succulent Manual: A guide to care and repair for all climates The Succulent Manual: A guide to care and repair for all climates by Andrea Afra Published by Sucs for You! sucsforyou.com © 2018 Andrea Afra All rights reserved. No portion of this book may be reproduced in any form without permission from the publisher, except as permitted by U.S. copyright law. For permission please contact: [email protected] This book is dedicated to the plants that have taught me to be patient with myself, my loved ones who have remained a steady source of light and faith, and to the wonderful friends I've made along this succulent journey. -

Greenhouse of UNI Del’S Greenhouse Joe and Joan Traylor Ben and Tina Donath Bev Edmondson Patricia Hampton
A special thank you to: Harry and Molly Stine and Stine Seeds Merle Philips The Shea Foundation Greenhouse of UNI Del’s Greenhouse Joe and Joan Traylor Ben and Tina Donath Bev Edmondson Patricia Hampton BUENA VISTA Iowa’s accessibly scaled, eye-opening university. Estelle Siebens Science Center 610 West Fourth Street Storm Lake, Iowa 50588 1 800 383 9600 ph www.bvu.edu Greenhouse Only in a greenhouse can you have a desert right next to a rainforest. The western most of the three rooms has a number of cacti, aloes, agaves and euphorbia collected from the American Southwest and South Africa. The middle room has many species from the warm and wet parts of our planet, several of which make good houseplants. The nearest room is reserved for research projects, new plants and display of plants that are blooming. Greenhouse funds were Rainforest provided by Stine Seeds. Bambusa verticillata (Gramineae) (Bamboo) Carissa grandiflora (Apocynaceae) (Natural Plum Jasmine) Cissus rhombifolia (Grape Ivy) Desert Citrus lemoni (Ritaceae) (Ponderosa Lemon) Adromischus cristatus (Crassulaceae)(Crinkle Leaf Plant) Cyperus alternifolius (Cyperaceae) Aloe brevifolia (Liliaceae) (Crocodile Jaws) Drypterus marginalis (Eastern Wood Fern) Astrophytum myriostigma (Cactaceae) (Bishop’s Cap) Evolvulus speciosa (Convulaceae) Bryophyllum daigremontianum (Crassulaceae) (Mother of thousands) Ficus benjamina (Braided Ficus Tree) Crassula arborescens (Crassulaceae) (Silver Dollar Jade) Ficus elastica (Rubber Plant) Crassula perforata (Crassulaceae) (String of Buttons) -

Reproductive Biology of Kalanchoe Laetivirens (Crassulaceae) in The
Research, Society and Development, v. 10, n. 1, e27010111567, 2021 (CC BY 4.0) | ISSN 2525-3409 | DOI: http://dx.doi.org/10.33448/rsd-v10i1.11567 Reproductive biology of Kalanchoe laetivirens (Crassulaceae) in the edaphoclimatic conditions of Santa Catarina, Brazil Biologia reprodutiva de Kalanchoe laetivirens (Crassulaceae) nas condições edafoclimáticas de Santa Catarina, Brasil Biología reproductiva de Kalanchoe laetivirens (Crassulaceae) en las condiciones edafoclimáticas de Santa Catarina, Brasil Received: 12/30/2020 | Reviewed: 01/08/2021 | Accept: 01/11/2021 | Published: 01/13/2021 Leonardo Norberto de Sousa Filho ORCID: https://orcid.org/0000-0002-6673-8214 Universidade Federal de Santa Catarina, Brazil E-mail: [email protected] Jean Bressan Albarello ORCID: https://orcid.org/0000-0001-8840-1893 Universidade Federal de Santa Catarina, Brazil E-mail: [email protected] Mayara Martins Cardozo ORCID: https://orcid.org/0000-0001-8506-2785 Universidade Federal de Santa Catarina, Brazil E-mail: [email protected] Márcia Regina Faita ORCID: https://orcid.org/0000-0003-1664-134X Universidade Federal de Santa Catarina, Brazil E-mail: [email protected] Cristina Magalhães Ribas dos Santos ORCID: https://orcid.org/0000-0002-9118-6730 Universidade Federal de Santa Catarina, Brazil E-mail: [email protected] Abstract Crassulaceae of the Kalanchoe laetivirens species have ornamental potential, but are poorly studied, especially their reproductive biology and trophic resources made available to visitors. The aim of this study, therefore, was to characterize the reproductive system of K. laetivirens through analyses of floral morphology, trophic resources and floral visitors. Floral structures, pollen / egg ratio, in vitro pollen germination, pollen grain structure in scanning microscopy and the availability of trophic resources by the evaluation of potential and instant nectar were described. -
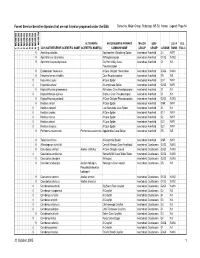
Sensitive Species That Are Not Listed Or Proposed Under the ESA Sorted By: Major Group, Subgroup, NS Sci
Forest Service Sensitive Species that are not listed or proposed under the ESA Sorted by: Major Group, Subgroup, NS Sci. Name; Legend: Page 94 REGION 10 REGION 1 REGION 2 REGION 3 REGION 4 REGION 5 REGION 6 REGION 8 REGION 9 ALTERNATE NATURESERVE PRIMARY MAJOR SUB- U.S. N U.S. 2005 NATURESERVE SCIENTIFIC NAME SCIENTIFIC NAME(S) COMMON NAME GROUP GROUP G RANK RANK ESA C 9 Anahita punctulata Southeastern Wandering Spider Invertebrate Arachnid G4 NNR 9 Apochthonius indianensis A Pseudoscorpion Invertebrate Arachnid G1G2 N1N2 9 Apochthonius paucispinosus Dry Fork Valley Cave Invertebrate Arachnid G1 N1 Pseudoscorpion 9 Erebomaster flavescens A Cave Obligate Harvestman Invertebrate Arachnid G3G4 N3N4 9 Hesperochernes mirabilis Cave Psuedoscorpion Invertebrate Arachnid G5 N5 8 Hypochilus coylei A Cave Spider Invertebrate Arachnid G3? NNR 8 Hypochilus sheari A Lampshade Spider Invertebrate Arachnid G2G3 NNR 9 Kleptochthonius griseomanus An Indiana Cave Pseudoscorpion Invertebrate Arachnid G1 N1 8 Kleptochthonius orpheus Orpheus Cave Pseudoscorpion Invertebrate Arachnid G1 N1 9 Kleptochthonius packardi A Cave Obligate Pseudoscorpion Invertebrate Arachnid G2G3 N2N3 9 Nesticus carteri A Cave Spider Invertebrate Arachnid GNR NNR 8 Nesticus cooperi Lost Nantahala Cave Spider Invertebrate Arachnid G1 N1 8 Nesticus crosbyi A Cave Spider Invertebrate Arachnid G1? NNR 8 Nesticus mimus A Cave Spider Invertebrate Arachnid G2 NNR 8 Nesticus sheari A Cave Spider Invertebrate Arachnid G2? NNR 8 Nesticus silvanus A Cave Spider Invertebrate Arachnid G2? NNR -

Forwarded Message
Volume 11, Number 4 October-November-December 1997 Nancy R. Morin and Judith M. Unger, Co-editors FLORA OF NORTH AMERICA NEWS The Flora of North America office has moved from the basement of the Administration Building—our home for about nine years—to the first floor of the new Monsanto Center at the Missouri Botanical Garden. We have graduated from frosted basement windows, where we could only tell if it was cloudy or sunny, to a south-facing wall of nothing but windows, with blinds to keep out too much sunlight. Come visit us in our new home. All of our phone numbers and our fax number have remained the same. The P.O. Box number is the same (299) as well as the zip code of 63166-0299, but our street address for package deliveries is 4500 Shaw Blvd. with a zip code of 63110. * * * * * Dr. Gerald Bane Straley (1945-1997) On 11 December 1997, we lost a friend and colleague, Gerald B. Straley, following a long and courageous battle. His continual optimism during his illness was an inspiration to all who knew him. Gerald was a Regional Coordinator, member of the Editorial Committee, and author for the FNA project. Gerald grew up on the family farm in Virginia and this undoubtedly initiated his great interest in nature. He completed his B.Sc. degree in Ornamental Horticulture at Virginia Polytechnic Institute and then went to Ohio University to study for his M.Sc. in Botany. In 1976, he came to do his Ph.D. on Arnica at the University of British Columbia under the guidance of Dr. -

Abstract Resumen
CARLOS VÁZQUEZ-COTERO1,3, VICTORIA SOSA1,2* PABLO CARRILLO-REYES3 Botanical Sciences 95 (3): 515-526, 2017 Abstract Background: Echeveria and Pachyphytum are two closely related Neotropical genera in the Crassulaceae. Several DOI: 10.17129/botsci.1190 species in Echeveria possess characters cited as diagnostic for Pachyphytum such as a clearly defned stem, a nectary scale on the inner face of petals and as inforescence a scorpioid cyme or cincinnus. Pachyphytum has been identifed as Copyright: © 2017 Vázquez-Cotero monophyletic while Echeveria as polyphyletic in previous molecular phylogenetic analysess. et al. This is an open access article distributed under the terms of the Hypothesis: The objective of this paper is to identify the phylogenetic position of a rare species with restricted distri- Creative Commons Attribution Li- bution in Echeveria, E. heterosepala that possesses the diagnostic characters of Pachyphytum to better understand the cense, which permits unrestricted generic limits between these two genera. We expect this species to be closely related to Pachyphytum. use, distribution, and reproduction Methods: Bayesian inference and Maximum Likelihood analyses were carried out using 47 taxa, including as ingroup, in any medium, provided the original species of Echeveria, Graptopetalum, Lenophyllum, Pachyphytum, Sedum, Thompsonella and Villadia and as outgroup, author and source are credited. species in Dudleya. Analyses were conducted based on plastid (rpl16, trnL-F) and nuclear (ETS, ITS) markers. Ances- tral character reconstruction was carried out under a parsimony criterion based on the molecular trees retrieved by the phylogenetic analyses. Four morphological characters were considered: defned stem, type of inforescence, nectary scale in petals and position of sepals. -

Plants for a Thirsty State
Plants for a Thirsty State by Nevin Smith and Ginny Hunt © 2016 Suncrest Nurseries, Inc. www.suncrestnurseries.com Introduction As we approach the close of a fourth critically dry season in California, it is time for those of us who love to garden to take stock of where we've been, and where we're going. It is probably safe to say that the days of extravagant water use are over, except perhaps in the wettest years. But a constructive challenge remains: How to continue to provide beauty and diversity in the garden with a more limited water budget. You may wonder whether there is really a good variety of water-conserving plants to choose from, in terms of forms, textures and colors, and whether they can be grown by ordinary mortals. This booklet is devoted to answering these questions with an emphatic "Yes". "Drought Tolerance" and the Real World Few phrases have been so much used and abused concerning garden plants in the past four years as "drought tolerant". The concept is well-intentioned, but it means radically different things to different people, including professional horticulturists. It can cover desert plants, like the agaves, that can thrive for months or years with no water beyond meager winter rains. It is more often applied to plants that will live, sometimes thrive, with natural winter rainfall in more temperate regions and irrigation every few weeks in the summer. And sometimes it is altogether misapplied, to plants that simply don't consume as much water as others for growth and maintenance, though they should never go dry. -

Molecular Phylogeny of the Acre Clade (Crassulaceae): Dealing with the Lack of Definitions for Echeveria and Sedum
Molecular Phylogenetics and Evolution 53 (2009) 267–276 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Molecular phylogeny of the Acre clade (Crassulaceae): Dealing with the lack of definitions for Echeveria and Sedum Pablo Carrillo-Reyes a,*, Victoria Sosa a, Mark E. Mort b a Departamento de Biología Evolutiva, Instituto de Ecología, A.C., Apartado Postal 63, 91070 Xalapa, Veracruz, Mexico b Department of Ecology and Evolutionary Biology and the Natural History Museum and Biodiversity Research Center, University of Kansas, 1200 Sunnyside Ave., Lawrence, KS 66045, USA article info abstract Article history: The phylogenetic relationships within many clades of the Crassulaceae are still uncertain, therefore in Received 24 February 2009 this study attention was focused on the ‘‘Acre clade”, a group comprised of approximately 526 species Revised 20 May 2009 in eight genera that include many Asian and Mediterranean species of Sedum and the majority of the Accepted 22 May 2009 American genera (Echeveria, Graptopetalum, Lenophyllum, Pachyphytum, Villadia, and Thompsonella). Par- Available online 29 May 2009 simony and Bayesian analyses were conducted with 133 species based on nuclear (ETS, ITS) and chloro- plast DNA regions (rpS16, matK). Our analyses retrieved four major clades within the Acre clade. Two of Keywords: these were in a grade and corresponded to Asian species of Sedum, the rest corresponded to a European– Altamiranoa Macaronesian group and to an American group. The American group included all taxa that were formerly ETS Graptopetalum placed in the Echeverioideae and the majority of the American Sedoideae. Our analyses support the ITS monophyly of three genera – Lenophyllum, Thompsonella, and Pachyphytum; however, the relationships Lenophyllum among Echeveria, Sedum and the various segregates of Sedum are largely unresolved. -

Linnaeus's Folly – Phylogeny, Evolution and Classification of Sedum
Messerschmid & al. • Phylogeny of Sedum and Sempervivoideae TAXON 69 (5) • October 2020: 892–926 SYSTEMATICS AND PHYLOGENY Linnaeus’s folly – phylogeny, evolution and classification of Sedum (Crassulaceae) and Crassulaceae subfamily Sempervivoideae Thibaud F.E. Messerschmid,1,2 Johannes T. Klein,3 Gudrun Kadereit2 & Joachim W. Kadereit1 1 Institut für Organismische und Molekulare Evolutionsbiologie, Johannes Gutenberg-Universität Mainz, 55099 Mainz, Germany 2 Institut für Molekulare Physiologie, Johannes Gutenberg-Universität Mainz, 55099 Mainz, Germany 3 Gothenburg Global Biodiversity Centre, Gothenburg, Sweden Address for correspondence: Thibaud Messerschmid, [email protected] DOI https://doi.org/10.1002/tax.12316 Abstract Sedum, containing approximately 470 species, is by far the largest genus of Crassulaceae. Three decades of molecular phy- logenetic work have provided evidence for the non-monophyly of Sedum and many more of the 30 genera of Crassulaceae subfam. Sempervivoideae. In this study, we present a broadly sampled and dated molecular phylogeny of Sempervivoideae including 80% of all infrageneric taxa described in Sedum as well as most other genera of the subfamily. We used sequences of one nuclear (ITS) and three plastid markers (matK, rps16, trnL-trnF). The five major lineages of Sempervivoideae (i.e., Telephium clade, Petrosedum clade, Sempervivum/Jovibarba, Aeonium clade, Leucosedum plus Acre clades) were resolved as successive sister to each other in the phylo- genetic analysis of the plastid markers, while in the ITS phylogeny the Petrosedum clade is the closest relative of the Aeonium clade. Our dating analysis of ITS suggests that Sempervivoideae diversified rapidly throughout the Paleocene and Eocene, possibly in the area of the former Tethys and Paratethys archipelago. -

Spring 2018 Plant List – 05/27/18 Miles' to Go - P.O
SPRING 2018 PLANT LIST – 05/27/18 MILES' TO GO - P.O. BOX 6 - CORTARO, AZ 85652 - 520-682-7272 - FAX 520-682-0480 EMAIL [email protected] - WEB SITE www.miles2go.com INFORMATION POLICY: We do not share or sell our mailing or emailing information with anyone. ORDERING INFORMATION Shipping Charges: Please use chart below to calculate shipping charges for your order. For orders under $25 - add $15.00 for shipping by Priority Mail For orders from $25 to $49.99 - add $10.00 for shipping by Priority Mail For orders $50 and over - free shipping by Priority Mail Express Mail will be billed for the exact amount, contact us if you are paying by check or money order. Catalog Number: Please include the catalog number which appears with each plant entry in order to speed up the processing of your order. Email Orders: At the current time we do not have a secured server and do not recommend emailing credit card numbers. If we have your credit card number on file, email us your order and include just the last 4 digits and exp. date of your credit card as verification. You may email your order and we will hold it for you until we receive payment information (via phone, fax or mail) . Fax Orders: You may fax your order to 520-682-0480 at any time. Foreign Orders: We cannot ship outside the United States and her territories. Minimum order: We have no minimum order but there is a shipping charge for some orders (See Shipping Charges.) Payment: We accept checks, money orders, Discover, Mastercard, Visa, and American Express. -

Andrea T. Kramer 2,3,4,5 , Jeremie B. Fant 3 , and Mary V. Ashley 4
American Journal of Botany 98(1): 109–121. 2011. I NFLUENCES OF LANDSCAPE AND POLLINATORS ON POPULATION GENETIC STRUCTURE: EXAMPLES FROM THREE PENSTEMON (PLANTAGINACEAE) SPECIES IN THE GREAT BASIN 1 Andrea T. Kramer 2,3,4,5 , Jeremie B. Fant 3 , and Mary V. Ashley 4 2 Botanic Gardens Conservation International (U.S.), 1000 Lake Cook Road, Glencoe, Illinois 60022 USA; 3 Chicago Botanic Garden, 1000 Lake Cook Road, Glencoe, Illinois 60022 USA; and 4 University of Illinois at Chicago, 845 West Taylor Street, M/C 066, Chicago, Illinois 60607 USA • Premise of the study : Despite rapid growth in the fi eld of landscape genetics, our understanding of how landscape features in- teract with life history traits to infl uence population genetic structure in plant species remains limited. Here, we identify popula- tion genetic divergence in three species of Penstemon (Plantaginaceae) similarly distributed throughout the Great Basin region of the western United States but with different pollination syndromes (bee and hummingbird). The Great Basin ’ s mountainous landscape provides an ideal setting to compare the interaction of landscape and dispersal ability in isolating populations of dif- ferent species. • Methods : We used eight highly polymorphic microsatellite loci to identify neutral population genetic structure between popula- tions within and among mountain ranges for eight populations of P. deustus , 10 populations of P. pachyphyllus , and 10 popula- tions of P. rostrifl orus . We applied traditional population genetics approaches as well as spatial and landscape genetics approaches to infer genetic structure and discontinuities among populations. • Key results : A ll three species had signifi cant genetic structure and exhibited isolation by distance, ranging from high structure and low inferred gene fl ow in the bee-pollinated species P. -

TURF REPLACEMENT PROGRAM MMWD LYL Approved Plant List
LANDSCAPE YOUR LAWN (LYL) TURF REPLACEMENT PROGRAM MMWD LYL Approved Plant List Attached is the current MMWD list of approved plants for the The values are obtained by determining the area of a circle using Landscape Your Lawn (LYL) Program. the plant spread or width as the diameter. To find the area of a circle, square the diameter and multiply by .7854. Squaring the This list is taken from the Water Use Classification of Landscape diameter means multiplying the diameter by itself. For example, a Species (WUCOLS IV) – a widely accepted and commonly used plant with a 5 foot spread would be calculated as follows: source of information on landscape plant water needs. Plants that .7854 x 5 ft diameter x 5 ft diameter = 20 sq ft (values are rounded are listed in WUCOLS IV as “low” or “very low” water use for the Bay to the nearest whole number). Area have been included on this list. However, plants that are considered invasive and are found on the MMWD Invasive Plant List For values not provided, please refer to reputable gardening books are not included in this list and will not be allowed for the LYL or nurseries in order to determine the diameter of the plant at program. maturity, or conduct an internet search using the botanical name and “mature size”. Any plants used in turf conversion that are not on this plant list will not count toward the 50 percent plant coverage requirement nor CA Natives will they be eligible for a rebate under LYL Option 1. Native plants are perfectly suited to our climate, soil, and animals.