Gutless Adenovirus: Last-Generation Adenovirus for Gene Therapy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mobile Genetic Elements in Streptococci
Curr. Issues Mol. Biol. (2019) 32: 123-166. DOI: https://dx.doi.org/10.21775/cimb.032.123 Mobile Genetic Elements in Streptococci Miao Lu#, Tao Gong#, Anqi Zhang, Boyu Tang, Jiamin Chen, Zhong Zhang, Yuqing Li*, Xuedong Zhou* State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, PR China. #Miao Lu and Tao Gong contributed equally to this work. *Address correspondence to: [email protected], [email protected] Abstract Streptococci are a group of Gram-positive bacteria belonging to the family Streptococcaceae, which are responsible of multiple diseases. Some of these species can cause invasive infection that may result in life-threatening illness. Moreover, antibiotic-resistant bacteria are considerably increasing, thus imposing a global consideration. One of the main causes of this resistance is the horizontal gene transfer (HGT), associated to gene transfer agents including transposons, integrons, plasmids and bacteriophages. These agents, which are called mobile genetic elements (MGEs), encode proteins able to mediate DNA movements. This review briefly describes MGEs in streptococci, focusing on their structure and properties related to HGT and antibiotic resistance. caister.com/cimb 123 Curr. Issues Mol. Biol. (2019) Vol. 32 Mobile Genetic Elements Lu et al Introduction Streptococci are a group of Gram-positive bacteria widely distributed across human and animals. Unlike the Staphylococcus species, streptococci are catalase negative and are subclassified into the three subspecies alpha, beta and gamma according to the partial, complete or absent hemolysis induced, respectively. The beta hemolytic streptococci species are further classified by the cell wall carbohydrate composition (Lancefield, 1933) and according to human diseases in Lancefield groups A, B, C and G. -

Viral Vectors and Biological Safety
Viral Vectors and Biological Safety Viral vectors are often designed so that they can enter human cells and deliver genes of interest. Viral vectors are usually replication-deficient – genes necessary for replication of the virus are removed from the vector and supplied separately through plasmids, helper virus, or packaging cell lines. There are several biosafety concerns that arise with the use of viral vectors including: 1) Tropism (host range) – viral vectors that can enter (infect) human cells are often used. 2) Replication-deficient viral vectors can gain back the deleted genes required for replication (become replication-competent) through recombination – referred to as replication-competent virus (RCV) breakthroughs. 3) Genes may be expressed in tissues and/or organisms where they are normally not expressed. In the case of some genes such as oncogenes, this could have far-reaching negative consequences. When evaluating safety for use of viral vectors, a number of factors need to be considered including: Risk Group (RG) of the organism; tropism (organism and tissue); route of transmission; whether the virus integrates into the host genome; and the specific gene(s) being introduced. Please contact the Office of Biological Safety (OBS) for more information on physical barriers and safety practices to use with specific viral vectors. This article concentrates on biological barriers that can be employed to improve safety when using viral vectors. Viral vectors frequently used are: • Retrovirus/lentivirus • Adenovirus • Adeno-associated virus (AAV) • Poxvirus • Herpes virus • Alphavirus • Baculovirus Amphotropic murine leukemia virus (MLV) – also called Moloney murine leukemia virus (MMLV) – and adenovirus are common viral vectors used to introduce genes into human cells. -
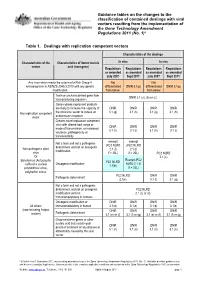
Guidance Tables on the Changes To
Guidance tables on the changes to the classification of contained dealings with viral vectors resulting from the implementation of the Gene Technology Amendment Regulations 2011 (No. 1)* Table 1. Dealings with replication competent vectors Characteristics of the dealings Characteristics of the Characteristics of donor nucleic In vitro In vivo vector acid (transgene) Regulations Regulations Regulations Regulations as amended as amended as amended as amended July 2007 Sept 2011* July 2007 Sept 2011* Any virus which meets the criteria of a Risk Group 4 Not Not microorganism in AS/NZS 2243.3:2010 with any genetic differentiated DNIR 3.1(p) differentiated DNIR 3.1(p) modification from below from below Toxin or uncharacterised gene from DNIR 3.1 (a), (b) or (c) toxin producing organism Genes whose expressed products are likely to increase the capacity of DNIR DNIR DNIR DNIR Any replication competent the virus/viral vector to induce an 3.1 (g) 3.1 (h) 3.1 (g) 3.1 (h) vector autoimmune response Creates novel replication competent virus with altered host range or DNIR DNIR DNIR DNIR mode of transmission, or increased 3.1 (h) 3.1 (i) 3.1 (h) 3.1 (i) virulence, pathogenicity or transmissibility exempt exempt Not a toxin and not a pathogenic (PC2 NLRD (PC2 NLRD determinant and not an oncogenic Non-pathogenic plant 2.1 (f) 2.1 (f) modification virus if > 25L) if > 25L) PC2 NLRD Or 2.1 (c) Exempt (PC2 Baculovirus (Autographa PC1 NLRD Oncogenic modification NLRD 2.1 (f) californica nuclear 1.1(b) polyhedrosis virus), if > 25L) polyhedrin minus PC2 NLRD -

Genetic Content and Evolution of Adenoviruses Andrew J
Journal of General Virology (2003), 84, 2895–2908 DOI 10.1099/vir.0.19497-0 Review Genetic content and evolution of adenoviruses Andrew J. Davison,1 Ma´ria Benko´´ 2 and Bala´zs Harrach2 Correspondence 1MRC Virology Unit, Institute of Virology, Church Street, Glasgow G11 5JR, UK Andrew Davison 2Veterinary Medical Research Institute, Hungarian Academy of Sciences, H-1581 Budapest, [email protected] Hungary This review provides an update of the genetic content, phylogeny and evolution of the family Adenoviridae. An appraisal of the condition of adenovirus genomics highlights the need to ensure that public sequence information is interpreted accurately. To this end, all complete genome sequences available have been reannotated. Adenoviruses fall into four recognized genera, plus possibly a fifth, which have apparently evolved with their vertebrate hosts, but have also engaged in a number of interspecies transmission events. Genes inherited by all modern adenoviruses from their common ancestor are located centrally in the genome and are involved in replication and packaging of viral DNA and formation and structure of the virion. Additional niche-specific genes have accumulated in each lineage, mostly near the genome termini. Capture and duplication of genes in the setting of a ‘leader–exon structure’, which results from widespread use of splicing, appear to have been central to adenovirus evolution. The antiquity of the pre-vertebrate lineages that ultimately gave rise to the Adenoviridae is illustrated by morphological similarities between adenoviruses and bacteriophages, and by use of a protein-primed DNA replication strategy by adenoviruses, certain bacteria and bacteriophages, and linear plasmids of fungi and plants. -

Human Adenovirus Encodes Two Proteins Which Have Opposite Effects on Accumulation of Alternatively Spliced Mrnas
MOLECULAR AND CELLULAR BIOLOGY, Jan. 1994, p. 437-445 Vol. 14, No. 1 0270-7306/94/$04.00+0 Copyright X 1994, American Society for Microbiology Human Adenovirus Encodes Two Proteins Which Have Opposite Effects on Accumulation of Alternatively Spliced mRNAs KATARINA NORDQVIST,t KARIN OHMAN, AND GORAN AKUSJARVI* Department of Cell and Molecular Biology, The Medical Nobel Institute, Karolinska Institutet, S-171 77 Stockholm, Sweden Received 23 June 1993/Returned for modification 17 August 1993/Accepted 30 September 1993 All mRNAs expressed from the adenovirus major late transcription unit have a common, 201-nucleotide-long 5' leader sequence, which consists of three short exons (the tripartite leader). This leader has two variants, either with or without the i-leader exon, which, when present, is spliced between the second and the third exons of the tripartite leader. Previous studies have shown that adenovirus early region 4 (E4) encodes two proteins, E4 open reading frame 3 (E4-ORF3) and E4-ORF6, which are required for efficient expression of mRNAs from the major late transcription unit. These two E4 proteins appear to have redundant activities, and expression of one has been shown to be sufficient for efficient maijor late mRNA accumulation during a lytic virus infection. In this report, we provide evidence that E4-ORF3 and E4-ORF6 both regulate major late mRNA accumulation by stimulating constitutive splicing. Moreover, we show that the two proteins have different effects on accumulation ofalternatively spliced tripartite leader exons. In a DNA transfection assay, E4-ORF3 was shown to facilitate i-leader exon inclusion, while E4-ORF6 preferentially favored i-leader exon skipping. -

THE ROLE of BOVINE ADENOVIRUS-3 PROTEIN V (Pv) in VIRUS REPLICATION
THE ROLE OF BOVINE ADENOVIRUS-3 PROTEIN V (pV) IN VIRUS REPLICATION A Thesis Submitted to the Faculty of Graduate Studies and Research in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Veterinary Microbiology University of Saskatchewan Saskatoon By Xin Zhao © Copyright Xin Zhao, June 2016. All rights reserved PERMISSION TO USE In presenting this thesis in partial fulfillment of the requirements for a postgraduate degree from the University of Saskatchewan, I agree that the libraries of this university may make it freely available for inspection. I further agree that permission for copying of this thesis in any manner, whole or in part, for scholarly purposes may be granted by the professors who supervised my thesis work or in their absence, the Head of the Department or the Dean of the college in which my thesis work was done. It is understood that any copying or publication or use of this thesis or parts thereof for financial gain shall not be allowed without any written permission. It is also understood that due recognition shall be given to me and to the University of Saskatchewan in any scholarly use which may be made of any material in my thesis. Request for permission to copy or to make other use of material in this thesis in whole or part should be addressed to: Head of the Department of Veterinary Microbiology University of Saskatchewan, Saskatoon, Saskatchewan, S7N 5B4 i ABSTRACT Bovine adenovirus type 3 (BAdV-3), which is a non-enveloped icosahedral particle with a double-stranded DNA genome of 34,446 base pair, has been developed as a vaccine vector. -

Adenovirus Gene Expression - a Model for Mammalian Cells
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Volume 74, number 2 FEBS LETTERS March 1977 ADENOVIRUS GENE EXPRESSION - A MODEL FOR MAMMALIAN CELLS Lennart PHILIPSON Department of Microbiology, Biomedical Center, Uppsala University S-751 23, Uppsala, Sweden Received 5 January 1977 Animal DNA viruses provide a powerful system for The synthesis of host cell RNA is unaffected at early analysis of transcription and translation in mammalian times but is suppressed late in productive infection. cells. During productive infection, the adenovirus DNA Only 1 O-20% of the normal amount of ribosomal enters the nucleus rapidly [ 1 ] and is transcribed into RNA is transported to the cytoplasm [9,16] late, large RNA molecules [2-41. Both early and late in although significant amounts of 45 S ribosomal productive infection nuclear RNA becomes polyadeny- precursor RNA appear to be synthesized [ 161. Late lated [5] and RNA molecules which are destined to after infection the synthesis of host-cell heterogeneous become messenger RNA (mRNA) are presumably nuclear RNA (HnRNA) is suppressed [17]. Nearly all cleaved before entering the cytoplasm [3, 6-91. A mRNA transported to the cytoplasm late after infection switch from early to late gene expression occurs at the is of viral origin. onset of viral DNA repliation and leads to quantitative The sequences of mRNA which are synthesized as well as qualitative changes in the synthesis of viral early after infection persist in the cytoplasm late after mRNA. Late after infection the cytoplasm contains infection although all early mRNA sequences are not about lo-times more viral RNA than at early times synthesized late after infection [2, 18-221. -

Adenovirus-Host Interactions: Implications for Tropism and Therapy
Adenovirus-host interactions: implications for tropism and therapy Annasara Lenman Department of Clinical Microbiology Umeå 2016 Responsible publisher under Swedish law: the Dean of the Medical Faculty This work is protected by the Swedish Copyright Legislation (Act 1960:729) ISBN: 978-91-7601-453-0 ISSN: 0346-6612-1798 Omslag gjort m.h.a. familj & vänner. Grafisk design av Lina Cabal ([email protected]). Elektronisk version tillgänglig på http://umu.diva-portal.org/ Tryck/Printed by: Print & Media Umeå, Sverige 2016 Till min älskade familj och mina underbara vänner Table of Contents TABLE OF CONTENTS .................................................................................. I ABSTRACT ................................................................................................... III SUMMARY IN SWEDISH-POPULÄRVETENSKAPLIG SAMMANFATTNING PÅ SVENSKA ............................................................ VI ABBREVIATIONS ....................................................................................... VIII LIST OF PAPERS .......................................................................................... X INTRODUCTION ............................................................................................ 1 HISTORY ....................................................................................................... 1 TAXONOMY ................................................................................................... 2 CLINICAL AND PATHOLOGICAL ASPECTS ......................................................... -

Viral Vectors
OCCUPATIONAL HEALTH CONSIDERATIONS FOR WORK WITH LENTIVIRAL VECTORS GARY R. FUJIMOTO, M.D. OCCUPATIONAL MEDICINE CONSULTANT OCTOBER 6, 2014 Disclosure: Lecture includes off-label use of antiretroviral medications VIRAL VECTORS Definition: Viruses engineered to deliver foreign genetic material (transgene) to cells Many viral vectors deliver the genetic material into the host cells but not into the host genome where the virus replicates (unless replication incompetent) Retroviral and lentiviral vectors deliver genetic transgenes into the host chromosomes LENTIVIRAL VECTORS Human immunodeficiency virus (HIV) is a lentivirus that infects both dividing and non-dividing cells Use of the HIV virus as a viral vector has required the reengineering of the virus to achieve safe gene transfer Since HIV normally targets CD4 cells, replacing the HIV envelope gene with vesicular stomatitis virus glycoprotein (VSV-G) expands the infectious range of the vector and modes of transmission LENTIVIRAL VECTORS Remember: replication deficient lentiviral vectors integrate the vector into the host chromosomes 3rd and 4th generation constructs unlikely to become replication competent with enhanced safety due to self- inactivating vectors (however, consider present or future HIV infection) Replication deficient lentiviral vectors should be regarded as single-event infectious agents Many researchers regard these agents as relatively benign although transgene integration does occur with generally unknown effects LENTIVIRAL OCCUPATIONAL EXPOSURES Lentiviral -

Viral Vectors
505 Oldham Court Lexington, KY 40502 Phone: (859) 257-1049 Fax: (859) 323-3838 E-Mail: [email protected] http://ehs.uky.edu/biosafety VIRAL VECTORS WHAT IS A VIRAL VECTOR? Viral vectors work like a “nanosyringe” to deliver nucleic acid to a target. They are often more efficient than other transfection methods, are useful for whole organism studies, have a relatively low toxicity, and are a likely route for human gene transfer. All viral vectors require a host for replication. The production of a viral vector is typically separated from the ability of the viral vector to infect cells. While viral vectors are not typically considered infectious agents, they do maintain their ability to “infect” cells. Viral vectors just don’t replicate (although there are some replicating viral vectors in use) under experimental conditions. An HIV- based lentiviral vector no longer possesses the ability to infect an individual with HIV, but it does maintain the ability to enter a cell and express genetic information. This is why viral vectors are useful, but also require caution. If a viral vector can transfect a human cell line on a plate, it can also transfect YOUR cells if accidentally exposed. SAFETY CONSIDERATIONS FOR ALL VIRAL VECTORS When utilizing ANY viral vector, the following questions must be addressed… 1. What potential does your method of viral vector production have to generate a replication competent virus? a. Generation of viral vector refers to the number of recombination events required to form a replication competent virus. For example, if you’re using a lentivirus that is split up between 4 plasmids (gag/pol, VSV-g, rev, transgene), 3 recombination events must take place to create a replication competent virus, therefore you are using a 3rd generation lentiviral vector. -

RNA Viruses As Tools in Gene Therapy and Vaccine Development
G C A T T A C G G C A T genes Review RNA Viruses as Tools in Gene Therapy and Vaccine Development Kenneth Lundstrom PanTherapeutics, Rte de Lavaux 49, CH1095 Lutry, Switzerland; [email protected]; Tel.: +41-79-776-6351 Received: 31 January 2019; Accepted: 21 February 2019; Published: 1 March 2019 Abstract: RNA viruses have been subjected to substantial engineering efforts to support gene therapy applications and vaccine development. Typically, retroviruses, lentiviruses, alphaviruses, flaviviruses rhabdoviruses, measles viruses, Newcastle disease viruses, and picornaviruses have been employed as expression vectors for treatment of various diseases including different types of cancers, hemophilia, and infectious diseases. Moreover, vaccination with viral vectors has evaluated immunogenicity against infectious agents and protection against challenges with pathogenic organisms. Several preclinical studies in animal models have confirmed both immune responses and protection against lethal challenges. Similarly, administration of RNA viral vectors in animals implanted with tumor xenografts resulted in tumor regression and prolonged survival, and in some cases complete tumor clearance. Based on preclinical results, clinical trials have been conducted to establish the safety of RNA virus delivery. Moreover, stem cell-based lentiviral therapy provided life-long production of factor VIII potentially generating a cure for hemophilia A. Several clinical trials on cancer patients have generated anti-tumor activity, prolonged survival, and -

Lentivirus and Lentiviral Vectors Fact Sheet
Lentivirus and Lentiviral Vectors Family: Retroviridae Genus: Lentivirus Enveloped Size: ~ 80 - 120 nm in diameter Genome: Two copies of positive-sense ssRNA inside a conical capsid Risk Group: 2 Lentivirus Characteristics Lentivirus (lente-, latin for “slow”) is a group of retroviruses characterized for a long incubation period. They are classified into five serogroups according to the vertebrate hosts they infect: bovine, equine, feline, ovine/caprine and primate. Some examples of lentiviruses are Human (HIV), Simian (SIV) and Feline (FIV) Immunodeficiency Viruses. Lentiviruses can deliver large amounts of genetic information into the DNA of host cells and can integrate in both dividing and non- dividing cells. The viral genome is passed onto daughter cells during division, making it one of the most efficient gene delivery vectors. Most lentiviral vectors are based on the Human Immunodeficiency Virus (HIV), which will be used as a model of lentiviral vector in this fact sheet. Structure of the HIV Virus The structure of HIV is different from that of other retroviruses. HIV is roughly spherical with a diameter of ~120 nm. HIV is composed of two copies of positive ssRNA that code for nine genes enclosed by a conical capsid containing 2,000 copies of the p24 protein. The ssRNA is tightly bound to nucleocapsid proteins, p7, and enzymes needed for the development of the virion: reverse transcriptase (RT), proteases (PR), ribonuclease and integrase (IN). A matrix composed of p17 surrounds the capsid ensuring the integrity of the virion. This, in turn, is surrounded by an envelope composed of two layers of phospholipids taken from the membrane of a human cell when a newly formed virus particle buds from the cell.