Viral Vectors Applied for Rnai-Based Antiviral Therapy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mobile Genetic Elements in Streptococci
Curr. Issues Mol. Biol. (2019) 32: 123-166. DOI: https://dx.doi.org/10.21775/cimb.032.123 Mobile Genetic Elements in Streptococci Miao Lu#, Tao Gong#, Anqi Zhang, Boyu Tang, Jiamin Chen, Zhong Zhang, Yuqing Li*, Xuedong Zhou* State Key Laboratory of Oral Diseases, National Clinical Research Center for Oral Diseases, West China Hospital of Stomatology, Sichuan University, Chengdu, PR China. #Miao Lu and Tao Gong contributed equally to this work. *Address correspondence to: [email protected], [email protected] Abstract Streptococci are a group of Gram-positive bacteria belonging to the family Streptococcaceae, which are responsible of multiple diseases. Some of these species can cause invasive infection that may result in life-threatening illness. Moreover, antibiotic-resistant bacteria are considerably increasing, thus imposing a global consideration. One of the main causes of this resistance is the horizontal gene transfer (HGT), associated to gene transfer agents including transposons, integrons, plasmids and bacteriophages. These agents, which are called mobile genetic elements (MGEs), encode proteins able to mediate DNA movements. This review briefly describes MGEs in streptococci, focusing on their structure and properties related to HGT and antibiotic resistance. caister.com/cimb 123 Curr. Issues Mol. Biol. (2019) Vol. 32 Mobile Genetic Elements Lu et al Introduction Streptococci are a group of Gram-positive bacteria widely distributed across human and animals. Unlike the Staphylococcus species, streptococci are catalase negative and are subclassified into the three subspecies alpha, beta and gamma according to the partial, complete or absent hemolysis induced, respectively. The beta hemolytic streptococci species are further classified by the cell wall carbohydrate composition (Lancefield, 1933) and according to human diseases in Lancefield groups A, B, C and G. -

Interactions De La Protéine Nsp1 Du Virus Chikungunya Avec Les Membranes De L’Hôte Et Conséquences Fonctionnelles William Bakhache
Interactions de la protéine nsP1 du virus Chikungunya avec les membranes de l’hôte et conséquences fonctionnelles William Bakhache To cite this version: William Bakhache. Interactions de la protéine nsP1 du virus Chikungunya avec les membranes de l’hôte et conséquences fonctionnelles. Médecine humaine et pathologie. Université Montpellier, 2020. Français. NNT : 2020MONTT008. tel-03132645 HAL Id: tel-03132645 https://tel.archives-ouvertes.fr/tel-03132645 Submitted on 5 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE POUR OBTENIR LE GRADE DE DOCTEUR DE L’UNIVERSITÉ DE M ONTPELLIER En Biologie Santé École Doctorale Sciences Chimiques et Biologiques pour la Santé UMR 9004 - Institut de Recherche en Infectiologie de Montpellier Interactions de la protéine nsP1 du virus Chikungunya avec les membranes de l’hôte et conséquences fonctionnelles Présentée par William Bakhache Le 27 Mars 2020 Sous la direction de Laurence Briant Devant le jury composé de Jean-Luc Battini, Directeur de Recherche, IRIM, Montpellier Président de jury Ali Amara, Directeur de recherche, U944 INSERM, Paris Rapporteur Juan Reguera, Chargé de recherche, AFMB, Marseille Rapporteur Raphaël Gaudin, Chargé de recherche, IRIM, Montpellier Examinateur Yves Rouillé, Directeur de Recherche, CIIL, Lille Examinateur Bruno Coutard, Professeur des universités, UVE, Marseille Co-encadrant de thèse Laurence Briant, Directeur de recherche, IRIM, Montpellier Directeur de thèse 1 Acknowledgments I want to start by thanking the members of my thesis jury. -

Gutless Adenovirus: Last-Generation Adenovirus for Gene Therapy
Gene Therapy (2005) 12, S18–S27 & 2005 Nature Publishing Group All rights reserved 0969-7128/05 $30.00 www.nature.com/gt CONFERENCE PAPER Gutless adenovirus: last-generation adenovirus for gene therapy R Alba1, A Bosch1 and M Chillon1,2 1Gene Therapy Laboratory, Department of Biochemistry and Molecular Biology, Center of Animal Biotechnology and Gene Therapy (CBATEG), Universitat Auto`noma de Barcelona, Bellaterra, Spain; and 2Institut Catala` de Recerca i Estudis Avanc¸ats (ICREA), Barcelona, Spain Last-generation adenovirus vectors, also called helper-depen- viral coding regions, gutless vectors require viral proteins dent or gutless adenovirus, are very attractive for gene therapy supplied in trans by a helper virus. To remove contamination because the associated in vivo immune response is highly by a helper virus from the final preparation, different systems reduced compared to first- and second-generation adenovirus based on the excision of the helper-packaging signal have vectors, while maintaining high transduction efficiency and been generated. Among them, Cre-loxP system is mostly tropism. Nowadays, gutless adenovirus is administered in used, although contamination levels still are 0.1–1% too high different organs, such as the liver, muscle or the central to be used in clinical trials. Recently developed strategies to nervous system achieving high-level and long-term transgene avoid/reduce helper contamination were reviewed. expression in rodents and primates. However, as devoid of all Gene Therapy (2005) 12, S18–S27. doi:10.1038/sj.gt.3302612 Keywords: adenovirus; gutless; helper-dependent vectors; in vivo gene therapy Introduction clinical for more information). Nowadays, adenovirus vectors are applied to treat cancer, monogenic disorders, Gene therapy for most genetic diseases requires expres- vascular diseases and others complications. -

Viral Strategies to Arrest Host Mrna Nuclear Export
Viruses 2013, 5, 1824-1849; doi:10.3390/v5071824 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Nuclear Imprisonment: Viral Strategies to Arrest Host mRNA Nuclear Export Sharon K. Kuss *, Miguel A. Mata, Liang Zhang and Beatriz M. A. Fontoura Department of Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; E-Mails: [email protected] (M.A.M.); [email protected] (L.Z.); [email protected] (B.M.A.F) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-214-633-2001; Fax: +1-214-648-5814. Received: 10 June 2013; in revised form: 27 June 2013 / Accepted: 11 July 2013 / Published: 18 July 2013 Abstract: Viruses possess many strategies to impair host cellular responses to infection. Nuclear export of host messenger RNAs (mRNA) that encode antiviral factors is critical for antiviral protein production and control of viral infections. Several viruses have evolved sophisticated strategies to inhibit nuclear export of host mRNAs, including targeting mRNA export factors and nucleoporins to compromise their roles in nucleo-cytoplasmic trafficking of cellular mRNA. Here, we present a review of research focused on suppression of host mRNA nuclear export by viruses, including influenza A virus and vesicular stomatitis virus, and the impact of this viral suppression on host antiviral responses. Keywords: virus; influenza virus; vesicular stomatitis virus; VSV; NS1; matrix protein; nuclear export; nucleo-cytoplasmic trafficking; mRNA export; NXF1; TAP; CRM1; Rae1 1. Introduction Nucleo-cytoplasmic trafficking of proteins and RNA is critical for proper cellular functions and survival. -

A Family of Mammalian E3 Ubiquitin Ligases That Contain the UBR Box Motif and Recognize N-Degrons Takafumi Tasaki,1 Lubbertus C
MOLECULAR AND CELLULAR BIOLOGY, Aug. 2005, p. 7120–7136 Vol. 25, No. 16 0270-7306/05/$08.00ϩ0 doi:10.1128/MCB.25.16.7120–7136.2005 Copyright © 2005, American Society for Microbiology. All Rights Reserved. A Family of Mammalian E3 Ubiquitin Ligases That Contain the UBR Box Motif and Recognize N-Degrons Takafumi Tasaki,1 Lubbertus C. F. Mulder,2 Akihiro Iwamatsu,3 Min Jae Lee,1 Ilia V. Davydov,4† Alexander Varshavsky,4 Mark Muesing,2 and Yong Tae Kwon1* Center for Pharmacogenetics and Department of Pharmaceutical Sciences, School of Pharmacy, University of Pittsburgh, Pittsburgh, Pennsylvania 152611; Aaron Diamond AIDS Research Center, The Rockefeller University, New York, New York 100162; Protein Research Network, Inc., Yokohama, Kanagawa 236-0004, Japan3; and Division of Biology, California Institute of Technology, Pasadena, California 911254 Received 15 March 2005/Returned for modification 27 April 2005/Accepted 13 May 2005 A subset of proteins targeted by the N-end rule pathway bear degradation signals called N-degrons, whose determinants include destabilizing N-terminal residues. Our previous work identified mouse UBR1 and UBR2 as E3 ubiquitin ligases that recognize N-degrons. Such E3s are called N-recognins. We report here that while double-mutant UBR1؊/؊ UBR2؊/؊ mice die as early embryos, the rescued UBR1؊/؊ UBR2؊/؊ fibroblasts still retain the N-end rule pathway, albeit of lower activity than that of wild-type fibroblasts. An affinity assay for proteins that bind to destabilizing N-terminal residues has identified, in addition to UBR1 and UBR2, a huge (570 kDa) mouse protein, termed UBR4, and also the 300-kDa UBR5, a previously characterized mammalian E3 known as EDD/hHYD. -

Viral Vectors and Biological Safety
Viral Vectors and Biological Safety Viral vectors are often designed so that they can enter human cells and deliver genes of interest. Viral vectors are usually replication-deficient – genes necessary for replication of the virus are removed from the vector and supplied separately through plasmids, helper virus, or packaging cell lines. There are several biosafety concerns that arise with the use of viral vectors including: 1) Tropism (host range) – viral vectors that can enter (infect) human cells are often used. 2) Replication-deficient viral vectors can gain back the deleted genes required for replication (become replication-competent) through recombination – referred to as replication-competent virus (RCV) breakthroughs. 3) Genes may be expressed in tissues and/or organisms where they are normally not expressed. In the case of some genes such as oncogenes, this could have far-reaching negative consequences. When evaluating safety for use of viral vectors, a number of factors need to be considered including: Risk Group (RG) of the organism; tropism (organism and tissue); route of transmission; whether the virus integrates into the host genome; and the specific gene(s) being introduced. Please contact the Office of Biological Safety (OBS) for more information on physical barriers and safety practices to use with specific viral vectors. This article concentrates on biological barriers that can be employed to improve safety when using viral vectors. Viral vectors frequently used are: • Retrovirus/lentivirus • Adenovirus • Adeno-associated virus (AAV) • Poxvirus • Herpes virus • Alphavirus • Baculovirus Amphotropic murine leukemia virus (MLV) – also called Moloney murine leukemia virus (MMLV) – and adenovirus are common viral vectors used to introduce genes into human cells. -
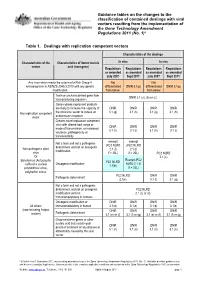
Guidance Tables on the Changes To
Guidance tables on the changes to the classification of contained dealings with viral vectors resulting from the implementation of the Gene Technology Amendment Regulations 2011 (No. 1)* Table 1. Dealings with replication competent vectors Characteristics of the dealings Characteristics of the Characteristics of donor nucleic In vitro In vivo vector acid (transgene) Regulations Regulations Regulations Regulations as amended as amended as amended as amended July 2007 Sept 2011* July 2007 Sept 2011* Any virus which meets the criteria of a Risk Group 4 Not Not microorganism in AS/NZS 2243.3:2010 with any genetic differentiated DNIR 3.1(p) differentiated DNIR 3.1(p) modification from below from below Toxin or uncharacterised gene from DNIR 3.1 (a), (b) or (c) toxin producing organism Genes whose expressed products are likely to increase the capacity of DNIR DNIR DNIR DNIR Any replication competent the virus/viral vector to induce an 3.1 (g) 3.1 (h) 3.1 (g) 3.1 (h) vector autoimmune response Creates novel replication competent virus with altered host range or DNIR DNIR DNIR DNIR mode of transmission, or increased 3.1 (h) 3.1 (i) 3.1 (h) 3.1 (i) virulence, pathogenicity or transmissibility exempt exempt Not a toxin and not a pathogenic (PC2 NLRD (PC2 NLRD determinant and not an oncogenic Non-pathogenic plant 2.1 (f) 2.1 (f) modification virus if > 25L) if > 25L) PC2 NLRD Or 2.1 (c) Exempt (PC2 Baculovirus (Autographa PC1 NLRD Oncogenic modification NLRD 2.1 (f) californica nuclear 1.1(b) polyhedrosis virus), if > 25L) polyhedrin minus PC2 NLRD -

Patenting Interfering RNA
Patenting Interfering RNA J. Douglas Schultz SPE Art Unit 1635 (571) 272-0763 [email protected] Oligonucleotide Inhibitors: Mechanisms of Action RNAi - Mechanism of Action • dsRNA induces sequence-specific degradation of homologous gene transcripts • RISC metabolizes dsRNA to small 21-23- nucleotide siRNAs – RISC contains dsRNase (“Dicer”), ssRNase (Argonaut 2 or Ago2) • RISC utilizes antisense strand as “guide” to find cleavable target siRNA Mechanism of Action miRNA Mechanism of Action Interfering RNA Glossary of Terms • RNAi – RNA interference • dsRNA – double stranded RNA • siRNA – small interfering RNA, double stranded, 21-23 nucleotides • shRNA – short hairpin RNA (doubled stranded by virtue of a ssRNA folding back on itself) • miRNA – micro RNA • RISC – RNA-induced silencing complex – Dicer – RNase endonuclease siRNA miRNA • Exogenously delivered • Endogenously produced • 21-23mer dsRNA • 21-23mer dsRNA • Acts through RISC • Acts through RISC • Induces homologous target • Induces homologous target cleavage cleavage • Perfect sequence match • Imperfect sequence match – Results in target degradation – Results in translation arrest RNAi Patentability issues Sample Claims: • A siRNA that inhibits expression of a nucleic acid encoding protein X. OR • A siRNA comprising a 2’-modification, wherein said modification comprises 2’-fluoro, 2’-O-methyl, or 2’- deoxy. (Note: no target recited) OR • A method of reducing tumor cell growth comprising administering siRNA targeting protein X. RNAi Patentability Issues 35 U.S.C. 101 – Utility • Credible/Specific/Substantial/Well Established. • Used to attempt modulation of gene expression in human diseases • Routinely investigate gene function in a high throughput fashion or to (see Rana RT, Nat. Rev. Mol. Cell Biol. 2007, Vol. 8:23-36). -

A Picorna-Like Virus Suppresses the N-End Rule Pathway to Inhibit Apoptosis
RESEARCH ARTICLE A picorna-like virus suppresses the N-end rule pathway to inhibit apoptosis Zhaowei Wang1,2†, Xiaoling Xia1,2,3†, Xueli Yang1, Xueyi Zhang1,2, Yongxiang Liu1,2, Di Wu1,2, Yuan Fang1,2, Yujie Liu1,2, Jiuyue Xu2, Yang Qiu2, Xi Zhou1,2* 1State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan, China; 2State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China; 3Guangzhou Key Laboratory of Insect Development Regulation and Application Research, Institute of Insect Science and Technology & School of Life Sciences, South China Normal University, Guangzhou, China Abstract The N-end rule pathway is an evolutionarily conserved proteolytic system that degrades proteins containing N-terminal degradation signals called N-degrons, and has emerged as a key regulator of various processes. Viruses manipulate diverse host pathways to facilitate viral replication and evade antiviral defenses. However, it remains unclear if viral infection has any impact on the N-end rule pathway. Here, using a picorna-like virus as a model, we found that viral infection promoted the accumulation of caspase-cleaved Drosophila inhibitor of apoptosis 1 (DIAP1) by inducing the degradation of N-terminal amidohydrolase 1 (NTAN1), a key N-end rule component that identifies N-degron to initiate the process. The virus-induced NTAN1 degradation is independent of polyubiquitylation but dependent on proteasome. Furthermore, the virus- induced N-end rule pathway suppression inhibits apoptosis and benefits viral replication. Thus, our findings demonstrate that a virus can suppress the N-end rule pathway, and uncover a new *For correspondence: mechanism for virus to evade apoptosis. -

A SARS-Cov-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing
A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing Supplementary Information Supplementary Discussion All SARS-CoV-2 protein and gene functions described in the subnetwork appendices, including the text below and the text found in the individual bait subnetworks, are based on the functions of homologous genes from other coronavirus species. These are mainly from SARS-CoV and MERS-CoV, but when available and applicable other related viruses were used to provide insight into function. The SARS-CoV-2 proteins and genes listed here were designed and researched based on the gene alignments provided by Chan et. al. 1 2020 . Though we are reasonably sure the genes here are well annotated, we want to note that not every protein has been verified to be expressed or functional during SARS-CoV-2 infections, either in vitro or in vivo. In an effort to be as comprehensive and transparent as possible, we are reporting the sub-networks of these functionally unverified proteins along with the other SARS-CoV-2 proteins. In such cases, we have made notes within the text below, and on the corresponding subnetwork figures, and would advise that more caution be taken when examining these proteins and their molecular interactions. Due to practical limits in our sample preparation and data collection process, we were unable to generate data for proteins corresponding to Nsp3, Orf7b, and Nsp16. Therefore these three genes have been left out of the following literature review of the SARS-CoV-2 proteins and the protein-protein interactions (PPIs) identified in this study. -

Depletion of Kcnq1ot1 Non-Coding RNA Does Not Affect Imprinting Maintenance in Stem Cells Michael C
DEVELOPMENT AND STEM CELLS RESEARCH ARTICLE 3667 Development 138, 3667-3678 (2011) doi:10.1242/dev.057778 © 2011. Published by The Company of Biologists Ltd Depletion of Kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells Michael C. Golding1,2,3,*,†, Lauren S. Magri1,2,3,†, Liyue Zhang2,3, Sarah A. Lalone1,2,3, Michael J. Higgins4 and Mellissa R. W. Mann1,2,3,‡ SUMMARY To understand the complex regulation of genomic imprinting it is important to determine how early embryos establish imprinted gene expression across large chromosomal domains. Long non-coding RNAs (ncRNAs) have been associated with the regulation of imprinting domains, yet their function remains undefined. Here, we investigated the mouse Kcnq1ot1 ncRNA and its role in imprinted gene regulation during preimplantation development by utilizing mouse embryonic and extra-embryonic stem cell models. Our findings demonstrate that the Kcnq1ot1 ncRNA extends 471 kb from the transcription start site. This is significant as it raises the possibility that transcription through downstream genes might play a role in their silencing, including Th, which we demonstrate possesses maternal-specific expression during early development. To distinguish between a functional role for the transcript and properties inherent to transcription of long ncRNAs, we employed RNA interference-based technology to deplete Kcnq1ot1 transcripts. We hypothesized that post-transcriptional depletion of Kcnq1ot1 ncRNA would lead to activation of normally maternal-specific protein-coding genes on the paternal chromosome. Post-transcriptional short hairpin RNA-mediated depletion in embryonic stem, trophoblast stem and extra-embryonic endoderm stem cells had no observable effect on the imprinted expression of genes within the domain, or on Kcnq1ot1 imprinting center DNA methylation, although a significant decrease in Kcnq1ot1 RNA signal volume in the nucleus was observed. -

(12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub
US 20040O86860A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0086860 A1 Sohail (43) Pub. Date: May 6, 2004 (54) METHODS OF PRODUCING RNAS OF Publication Classification DEFINED LENGTH AND SEQUENCE (51) Int. Cl." .............................. C12Q 1/68; C12P 19/34 (76) Inventor: Muhammad Sohail, Marston (GB) (52) U.S. Cl. ............................................... 435/6; 435/91.2 Correspondence Address: MINTZ, LEVIN, COHN, FERRIS, GLOWSKY (57) ABSTRACT AND POPEO, PC. ONE FINANCIAL CENTER Methods of making RNA duplexes and single-stranded BOSTON, MA 02111 (US) RNAS of a desired length and Sequence based on cleavage of RNA molecules at a defined position, most preferably (21) Appl. No.: 10/264,748 with the use of deoxyribozymes. Novel deoxyribozymes capable of cleaving RNAS including a leader Sequence at a (22) Filed: Oct. 4, 2002 Site 3' to the leader Sequence are also described. Patent Application Publication May 6, 2004 Sheet 1 of 2 US 2004/0086860 A1 DNA Oligonucleotides T7 Promoter -TN-- OR 2N-2-N-to y Transcription Products GGGCGAAT-N-UU GGGCGAAT-N-UU w N Deoxyribozyme Cleavage - Q GGGCGAAT -------' Racction GGGCGAAT N-- UU N-UU ssRNA products N-UU Anneal ssRNA UU S-2N- UU siRNA product FIGURE 1: Flowchart summarising the procedure for siRNA synthesis. Patent Application Publication May 6, 2004 Sheet 2 of 2 US 2004/0086860 A1 Full-length transcript 3'-digestion product 5'-digestion product (5'GGGCGAATA) A: Production of single-stranded RNA templates by in vitro transcription and digestion With a deoxyribozyme V 2- 2 V 22inv 22 * 2 &3 S/AS - 88.8x, *...* or as IGFR -- is as 4.