The Origin, Evolution and Conservation of the Arran Sorb Us Microspecies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

List of Vascular Plants Endemic to Britain, Ireland and the Channel Islands 2020
British & Irish Botany 2(3): 169-189, 2020 List of vascular plants endemic to Britain, Ireland and the Channel Islands 2020 Timothy C.G. Rich Cardiff, U.K. Corresponding author: Tim Rich: [email protected] This pdf constitutes the Version of Record published on 31st August 2020 Abstract A list of 804 plants endemic to Britain, Ireland and the Channel Islands is broken down by country. There are 659 taxa endemic to Britain, 20 to Ireland and three to the Channel Islands. There are 25 endemic sexual species and 26 sexual subspecies, the remainder are mostly critical apomictic taxa. Fifteen endemics (2%) are certainly or probably extinct in the wild. Keywords: England; Northern Ireland; Republic of Ireland; Scotland; Wales. Introduction This note provides a list of vascular plants endemic to Britain, Ireland and the Channel Islands, updating the lists in Rich et al. (1999), Dines (2008), Stroh et al. (2014) and Wyse Jackson et al. (2016). The list includes endemics of subspecific rank or above, but excludes infraspecific taxa of lower rank and hybrids (for the latter, see Stace et al., 2015). There are, of course, different taxonomic views on some of the taxa included. Nomenclature, taxonomic rank and endemic status follows Stace (2019), except for Hieracium (Sell & Murrell, 2006; McCosh & Rich, 2018), Ranunculus auricomus group (A. C. Leslie in Sell & Murrell, 2018), Rubus (Edees & Newton, 1988; Newton & Randall, 2004; Kurtto & Weber, 2009; Kurtto et al. 2010, and recent papers), Taraxacum (Dudman & Richards, 1997; Kirschner & Štepànek, 1998 and recent papers) and Ulmus (Sell & Murrell, 2018). Ulmus is included with some reservations, as many taxa are largely vegetative clones which may occasionally reproduce sexually and hence may not merit species status (cf. -

Conserving Europe's Threatened Plants
Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation By Suzanne Sharrock and Meirion Jones May 2009 Recommended citation: Sharrock, S. and Jones, M., 2009. Conserving Europe’s threatened plants: Progress towards Target 8 of the Global Strategy for Plant Conservation Botanic Gardens Conservation International, Richmond, UK ISBN 978-1-905164-30-1 Published by Botanic Gardens Conservation International Descanso House, 199 Kew Road, Richmond, Surrey, TW9 3BW, UK Design: John Morgan, [email protected] Acknowledgements The work of establishing a consolidated list of threatened Photo credits European plants was first initiated by Hugh Synge who developed the original database on which this report is based. All images are credited to BGCI with the exceptions of: We are most grateful to Hugh for providing this database to page 5, Nikos Krigas; page 8. Christophe Libert; page 10, BGCI and advising on further development of the list. The Pawel Kos; page 12 (upper), Nikos Krigas; page 14: James exacting task of inputting data from national Red Lists was Hitchmough; page 16 (lower), Jože Bavcon; page 17 (upper), carried out by Chris Cockel and without his dedicated work, the Nkos Krigas; page 20 (upper), Anca Sarbu; page 21, Nikos list would not have been completed. Thank you for your efforts Krigas; page 22 (upper) Simon Williams; page 22 (lower), RBG Chris. We are grateful to all the members of the European Kew; page 23 (upper), Jo Packet; page 23 (lower), Sandrine Botanic Gardens Consortium and other colleagues from Europe Godefroid; page 24 (upper) Jože Bavcon; page 24 (lower), Frank who provided essential advice, guidance and supplementary Scumacher; page 25 (upper) Michael Burkart; page 25, (lower) information on the species included in the database. -

NSA Special Qualities
Extract from: Scottish Natural Heritage (2010). The special qualities of the National Scenic Areas . SNH Commissioned Report No.374. The Special Qualities of the North Arran National Scenic Area • A mountain presence that dominates the Firth of Clyde • The contrast between the wild highland interior and the populated coastal strip • The historical landscape in miniature • A dramatic, compact mountain area • A distinctive coastline with a rich variety of forms • One of the most important geological areas in Britain • An exceptional area for outdoor recreation • The experience of highland and island wildlife at close hand Special Quality Further information • A mountain presence that dominates the Firth of Clyde Soaring above the sea, the Arran Arran has been described as ‘the sleeping giant’, its head mountains with their distinctive profile and body comprising the mountains and moors of the island. hold the eye and dominate the Firth of Clyde and its surrounds. Sometimes they The peaks of the NSA can be seen from along the North are clear and distinct, reflected in a Ayrshire coast and from many places inland. It likewise mirror-calm sea, at other times they are dominates the views from the east coast of the Kintyre capped in cloud or wreathed in mist. peninsula, and there are further imposing views from Bute. Gemmell (1998) states that, because the presence of the island is so dominating, the Firth of Clyde cannot be imagined without the presence of Arran and its mountains. • The contrast between the wild highland interior and the populated coastal strip The contrast between the upland and The highland interior of North Arran is comparable with lowland landscapes is striking. -

Sorbus Pseudomeinichii - Evolution, Extinction and an Exciting Discovery on the Isle of Arran
Sorbus pseudomeinichii - Evolution, extinction and an exciting discovery on the Isle of Arran by R. Blackhall-Miles and Ben Ram, www.fossilplants.co.uk Evolution of Sorbus pseudomeinichii Ashley Robertson Timeline of discovery for x Sorbus pseudomeinichii 1901 Johan Hedlund publishes details of two new Sorbus species: S. meinichii from Norway and S. arranensis from Arran, Scotland. Sorbus aucuparia L. Sorbus rupicola (Syme) Hedl. 1957 Edmund Warburg describes a second species unique to Arran, Sorbus x pseudofennica; a backcross between S. aucuparia and S. arranensis 1949 Donald McVean collected a Sorbus in Glen Catacol, it is misidentified as S. arranensis. 1996 Phil Lusby saw McVean’s specimen and suggests it is a hybrid between Sorbus aucuparia Sorbus arranensis Hedl. S. aucuparia and S. pseudofennica. x 2000 Ashley Robertson re-finds tree and two others like it and proves the specimens, through genetic research, to be a back cross between S. aucuparia and S. pseudofennica. 2004 Searches fail to re-find the two additional trees; one had been killed in a Sorbus aucuparia Sorbus pseudofennica E. F. Warb flood and the other presumed to be eaten by deer. 2006 Ashley Robertson publishes Sorbus pseudomeinichii as a new species naming it for its resemblance to Hedlund’s S. meinichii it instantly becomes one of the rarest trees in the world. 2017 Sorbus pseudomeinichii published as Critically Endangered on the IUCN Red List (www.iucnredlist.orgspecies/79749367/79749371) A leaf from the newly discovered Sorbus pseudomeinichii plant of Sorbus pseudomeinichii 2020 - A special find On the 15th September, whilst visiting Arran to see it’s endemic Sorbus microspecies, two lone, unprotected and heavily grazed Sorbus trees were observed by us near the top of the Catacol burn. -
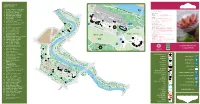
The-Commonwealth-Collection.Pdf
Round Border COMMONWEALTH Fern COLLECTION Garden Gate River W 1. Protea cyanoides - South Africa alk 2. Strelitzia reginae - South Africa GENERAL INFORMATION 3. Acacia karroo - Zambia Long Pit Glasshouse OPENING HOURS 4. Banksia ericifolia - Australia 32 GROUNDS 7 am to dusk (all year) 5. Wollemia nobilis - Australia 33 31 GLASSHOUSES 10 am - 6 pm (4.15 pm in winter) 30 6. Leptospermum scoparium - CHARGES 29 stairs New Zealand Hopkirk Glasgow City Council maintains a policy of free entry Building 7. Cyathea arborea - Jamaica 28 THE ENQUIRIES OFFICE: is situated behind the main range 27 25 15 and further information may be obtained there. 11 Enquiries 8. Ficus carica - Malta 26 24 23 20 21 22 10 19 Office PHONE: 0141 276 1614 9. Phormium tenax - New Zealand 17 Main Range FAX: 0141 276 1615 18 Glasshouses Curator’s 10. Angraecum podochiloides - 16 14 EMAIL: [email protected] House WEB: glasgowbotanicgardens.com or 13 Private Cameroon 12 www.glasgow.gov.uk/parks 11. Ansellia africana - Zambia 9 GROUPS VISITS: are especially welcome and a guide may be 12. Bowiea volubilis - Kenya Botanic Gardens available if arranged in advance with the Gardens’ Office. 13. Cycas seemannii - Fiji Tea Room DOGS: are allowed in the grounds, but should be kept on a 14. Ficus benjamina - India short leash. Dogs are not permitted in the glasshouses with the Peter exception of guide dogs. 15. Heliconia psittacorum - Walker TRANSPORT: by bus from the City Centre nos. 6, 6a, 6b, Trinidad & Tobago Memorial QUEEN 8, 8a, 10a, and 19. 16. Elettaria cardamomum - Sri Lanka MARGARET By Underground - to Hillhead Station 17. -

Inland Rock Outcrop and Scree Habitats (Uk Bap Priority Habitat)
INLAND ROCK OUTCROP AND SCREE HABITATS (UK BAP PRIORITY HABITAT) Summary This priority type includes plant communities that are confined or almost confined to inaccessible ledges on cliffs and crags or to screes and boulders. The predominant controlling factor is the base-status of the rock which determines what the vegetation consists of. There are communities of smaller ferns on small ledges, and in crevices, on screes and boulder-fields, and boulder-scree on sheltered slopes where snow lies late in spring. The habitat is common throughout the uplands where extensive glaciation has resulted in steep cliffs and outcrops, screes and block litter, though there are also examples on river cliffs in the lowlands, and the small fern communities can occur on walls and buildings. This priority habitat occurs throughout Scotland from just above sea level to over 1000 m on our highest hills. Rock and scree habitats are home to many rare and uncommon species. Boulder-fields, screes and high ledges where snow lies very late are habitats for snow-tolerant species. This priority habitat is also very important for lichens which are often the dominant life-form. Inland crags are the preferred nesting sites of golden eagle Aquila chrysaetos, sea eagle Haliaeetus albicilla, raven Corvus corax and peregrine falcon Falco peregrinus, whilst snow buntings Plectrophenax nivalis nest among boulders in high corries. The vegetation is near- natural. Being out of the reach of all grazing animals apart from goats, and being difficult to burn, these communities are not threatened by many human activities apart from some disturbance by climbers. -

Lead Ecosystem Group Code SBL Habitat Name UKBAP Priority
Lead Ecosystem Group code SBL Habitat name UKBAP Priority habitat name Category M&C H7 Calluna vulgaris-Scilla verna heath Maritime cliff and slopes 3 F&L CG2 Festuca ovina-Helictotrichon pratense grassland Lowland calcareous grassland 1 F&L CG7 Festuca ovina-Hieracium pilosella-Thymus polytrichus grassland Lowland calcareous grassland 1 M&C H7 Calluna vulgaris-Scilla verna heath Maritime cliff and slopes 3 F&L H8 Calluna vulgaris-Ulex gallii heath Lowland heathland 2 F&W M13 Schoenus nigricans-Juncus subnodulosus mire Lowland fens 1 F&L M13 Schoenus nigricans-Juncus subnodulosus mire Upland flushes, fens and swamps 3 F&L M21 Narthecium ossifragum-Sphagnum papillosum valley mire Upland flushes, fens and swamps 3 F&L M23 Juncus effusus/acutiflorus-Galium palustre rush-pasture Upland flushes, fens and swamps 3 F&L M23 Juncus effusus/acutiflorus-Galium palustre rush-pasture Purple moor grass and rush pastures 2 F&W M23 Juncus effusus/acutiflorus-Galium palustre rush-pasture Lowland fens 1 F&L M26 Molinia caerulea-Crepis paludosa fen Purple moor grass and rush pastures 2 F&W M26 Molinia caerulea-Crepis paludosa fen Lowland fens 1 F&W MG11 Festuca rubra-Agrostis stolonifera-Potentilla anserina inundation grassland Coastal and floodplain grazing marsh 3 M&C MG11 Festuca rubra-Agrostis stolonifera-Potentilla anserina inundation grassland Coastal saltmarsh 3 F&L MG11 Festuca rubra-Agrostis stolonifera-Potentilla anserina inundation grassland Open mosaic habitats on previously developed land 3 F&W MG12 Festuca arundinacea coarse grassland Coastal -

OPP DOC.19.21 Current OMEGA WEST RAW DATA
Total Taxon group Common name Scientific name Designation code Designation group 0 LICHEN Buellia hyperbolica Buellia hyperbolica IUCN Global Red List - Vulnerable, Nationally Rare, NERC S41, UK BAP Priority Species European/National Importance,European and UK Legal Protection 0 LICHEN Lecidea mucosa Lecidea mucosa Nationally Rare European/National Importance 0 FLOWERING PLANT Keeled Garlic Allium carinatum Invasive Non-Native Species Invasive Non-Native 0 LICHEN Micarea submilliaria Micarea submilliaria Nationally Rare European/National Importance 0 CHROMIST Macrocystis pyrifera Macrocystis pyrifera Wildlife and Countryside Act Schedule 9 European and UK Legal Protection 0 CHROMIST Macrocystis laevis Macrocystis laevis Wildlife and Countryside Act Schedule 9 European and UK Legal Protection 0 FLOWERING PLANT Indian Balsam Impatiens glandulifera Invasive Non-Native Species, Wildlife and Countryside Act Schedule 9 Invasive Non-Native,European and UK Legal Protection 0 FLOWERING PLANT False-acacia Robinia pseudoacacia Invasive Non-Native Species, Wildlife and Countryside Act Schedule 9 Invasive Non-Native,European and UK Legal Protection 0 FLOWERING PLANT Giant Hogweed Heracleum mantegazzianum Invasive Non-Native Species, Wildlife and Countryside Act Schedule 9 Invasive Non-Native,European and UK Legal Protection 0 CHROMIST Macrocystis integrifolius Macrocystis integrifolius Wildlife and Countryside Act Schedule 9 European and UK Legal Protection 0 CHROMIST Macrocystis augustifolius Macrocystis augustifolius Wildlife and Countryside Act -

Scottish Biodiversity List: Ferns & Flowering Plants
Scottish Biodiversity List: Ferns & Flowering Plants Taxonomic Scientific Name Common Name Distribution Group Map fern Asplenium obovatum Lanceolate Spleenwort NBNAtlas lanceolatum fern Cystopteris dickieana Dickie's Bladder-fern NBNAtlas fern Cystopteris montana Mountain Bladder-fern NBNAtlas fern Diphasiastrum complanatum Issler's Clubmoss NBNAtlas fern Gymnocarpium robertianum Limestone Fern NBNAtlas fern Lycopodiella inundata Marsh Clubmoss NBNAtlas fern Pilularia globulifera Pillwort NBNAtlas fern Polystichum lonchitis Holly-fern NBNAtlas fern Thelypteris palustris Marsh Fern NBNAtlas fern Trichomanes speciosum Killarney Fern NBNAtlas fern Woodsia alpina Alpine Woodsia NBNAtlas fern Woodsia ilvensis Oblong Woodsia NBNAtlas flowering plant Ajuga pyramidalis Pyramidal Bugle NBNAtlas flowering plant Alchemilla glaucescens Lady's Mantle NBNAtlas flowering plant Allium oleraceum Field Garlic NBNAtlas flowering plant Alopecurus myosuroides Black-grass NBNAtlas flowering plant Anagallis arvensis Scarlet Pimpernel NBNAtlas flowering plant Apium graveolens Wild Celery NBNAtlas flowering plant Arabis alpina Alpine Rock-cress NBNAtlas flowering plant Arenaria norvegica Arctic Sandwort NBNAtlas flowering plant Artemisia norvegica Norwegian Mugwort NBNAtlas flowering plant Astragalus alpinus Alpine Milk-vetch NBNAtlas flowering plant Astragalus danicus Purple Milk-vetch NBNAtlas flowering plant Bartsia alpina Alpine Bartsia NBNAtlas flowering plant Blysmus compressus Flat-sedge NBNAtlas flowering plant Brassica oleracea Wild Cabbage NBNAtlas -

6. Survey and Monitoring
6. Survey and monitoring Survey and monitoring are essential components of good Surveying management. Surveys of the scrub itself give information on extent, species composition and structure, and are essential ‘Survey’ is the recording of qualitative or quantitative for planning management. By using standard, repeatable biological data using easily repeatable standardised techniques for the initial surveys, they become the techniques over a restricted period without preconception baseline against which further monitoring is done. of the results. Surveys of species associated with scrub provide ‘Monitoring’ is the comparison of repeated surveys. It is information on their distribution and status, which is critically important that initial (baseline) surveys are done essential when planning management. Many scrub to a standard, described method and that the results are dependent species are now rare, due to loss and fully documented so that they can be repeated. fragmentation of their preferred scrub habitat. Management decisions made without regard to rare Baseline information should be gathered to inform species could damage or extinguish them; for example, management decisions and ongoing monitoring is needed eradication of willow scrub to prevent succession on a to continuously refine management techniques. This is wet heath could cause the local loss of the rare Dingy especially important on sites designated for nature Mocha moth. conservation. In England and Wales, targets have been set to ensure that SSSIs are in, or moving towards, It is theoretically possible to survey everything within the favourable condition. It is the responsibility of the statutory scrub community but this would take a great deal of time nature conservation body to assess whether this is the and money. -

The Vascular Plant Red Data List for Great Britain
Species Status No. 7 The Vascular Plant Red Data List for Great Britain Christine M. Cheffings and Lynne Farrell (Eds) T.D. Dines, R.A. Jones, S.J. Leach, D.R. McKean, D.A. Pearman, C.D. Preston, F.J. Rumsey, I.Taylor Further information on the JNCC Species Status project can be obtained from the Joint Nature Conservation Committee website at http://www.jncc.gov.uk/ Copyright JNCC 2005 ISSN 1473-0154 (Online) Membership of the Working Group Botanists from different organisations throughout Britain and N. Ireland were contacted in January 2003 and asked whether they would like to participate in the Working Group to produce a new Red List. The core Working Group, from the first meeting held in February 2003, consisted of botanists in Britain who had a good working knowledge of the British and Irish flora and could commit their time and effort towards the two-year project. Other botanists who had expressed an interest but who had limited time available were consulted on an appropriate basis. Chris Cheffings (Secretariat to group, Joint Nature Conservation Committee) Trevor Dines (Plantlife International) Lynne Farrell (Chair of group, Scottish Natural Heritage) Andy Jones (Countryside Council for Wales) Simon Leach (English Nature) Douglas McKean (Royal Botanic Garden Edinburgh) David Pearman (Botanical Society of the British Isles) Chris Preston (Biological Records Centre within the Centre for Ecology and Hydrology) Fred Rumsey (Natural History Museum) Ian Taylor (English Nature) This publication should be cited as: Cheffings, C.M. & Farrell, L. (Eds), Dines, T.D., Jones, R.A., Leach, S.J., McKean, D.R., Pearman, D.A., Preston, C.D., Rumsey, F.J., Taylor, I. -

Antibacterial Activity of Medicinal Plants from the Physicians of Myddvai, a 13Th Century Welsh Medical Manuscript
Author’s Accepted Manuscript Antibacterial activity of medicinal plants from The Physicians of Myddvai, a 13th century Welsh medical manuscript Charles Wagner, Jillian De Gezelle, Maureen Robertson, Keith Robertson, Mickey Wilson, Slavko Komarnytsky www.elsevier.com/locate/jep PII: S0378-8741(16)32508-9 DOI: http://dx.doi.org/10.1016/j.jep.2017.03.039 Reference: JEP10788 To appear in: Journal of Ethnopharmacology Received date: 22 December 2016 Revised date: 22 March 2017 Accepted date: 23 March 2017 Cite this article as: Charles Wagner, Jillian De Gezelle, Maureen Robertson, Keith Robertson, Mickey Wilson and Slavko Komarnytsky, Antibacterial activity of medicinal plants from The Physicians of Myddvai, a 13th century Welsh medical manuscript, Journal of Ethnopharmacology, http://dx.doi.org/10.1016/j.jep.2017.03.039 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting galley proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. Antibacterial activity of medicinal plants from The Physicians of Myddvai , a 13 th century Welsh medical manuscript Charles Wagner 1,2,3 , Jillian De Gezelle 2, Maureen Robertson 3, Keith Robertson 3, Mickey Wilson 1, Slavko Komarnytsky 1,2,4* 1Plants