Freestyle 1.0 User Guide Version A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Computer Essentials – Session 1 – Step-By-Step Guide
Computer Essentials – Session 1 – Step-by-Step Guide Note: Completing the Mouse Tutorial and Mousercise exercise which are available on the Class Resources webpage constitutes the first part of this lesson. ABOUT PROGRAMS AND OPERATING SYSTEMS Any time a task is performed on a computer, it is done through a program. For the user, the program is the application – the tool – for accomplishing a task. For the computer, it is a set of instructions on knowing how to perform this task. Examples of programs include Internet Explorer and Microsoft Word. The most important program overall is the operating system, which manages all of the computer’s resources and decides how to treat input from the user. There are several different operating systems in circulation, such as Mac O/S and Linux. However, far and away, the most commonly-used operating system is Microsoft Windows. (Note that Microsoft is just the name of the company that makes Windows, and that there are several versions of Windows. In 2012 all the computers in the lab were updated to the version of Windows called Windows 7. If you have some experience with the older versions of Windows you will notice that things look a bit different on a computer running Windows 7. THE WINDOWS 7 ENVIRONMENT The Desktop The first thing you see when you log on to the computer is the DESKTOP. The Desktop is the display area you see when Windows opens. The following items are the most common items that can be found on the desktop: Mouse pointer Icons Shortcuts to Storage drives Notification Start Button Taskbar tray Show Desktop/Peek button Andrea Philo September 2012 Page 1 of 13 Montgomery County-Norristown Public Library Computer Essentials – Session 1 – Step-by-Step Guide Parts of the Windows 7 Desktop Icon: A picture representing a program or file or places to store files. -

Adding the Maine Prescription Monitoring System As a Favorite
Adding the Maine Prescription Monitoring System as a Favorite Beginning 1/1/17 all newly prescribed narcotic medications ordered when discharging a patient from the Emergency Department or an Inpatient department must be recorded in the Maine prescription monitoring system (Maine PMP). Follow these steps to access the Maine PMP site from the SeHR/Epic system and create shortcuts to the activity to simplify access. Access the Maine PMP site from hyperspace 1. Follow this path for the intial launch of the Maine PMP site: Epic>Help>Maine PMP Program Please note: the Maine PMP database is managed and maintained by the State of Maine and does NOT support single-sign on 2. Click Login to enter your Maine PMP user name and password a. If you know your user name and password, click Login b. To retrieve your user name click Retrieve User Name c. To retrieve your password click Retrieve Password Save the Maine PMP site as a favorite under the Epic button After accessing the site from the Epic menu path you can make the link a favorite under the Epic menu. 1. Click the Epic button, the Maine PMP program hyperlink will appear under your Recent activities, click the Star icon to save it as a favorite. After staring the activity it becomes a favorite stored under the Epic button Save the Maine PMP site as a Quick Button on your Epic Toolbar After accessing the site from the Epic menu path you may choose to make the Maine PMP hyperlink a quick button on your main Epic toolbar. -

Basic Computer Lesson
Table of Contents MICROSOFT WORD 1 ONE LINC What is MSWord? MSWord is a word-processing program that allows users to create, edit, and enhance text in a variety of formats. Word is a powerful word processor with sophisticated editing and formatting as well as graphic- enhancement capabilities. Word is a good program for novice users since it is relatively easy to learn and can be integrated with language learning. Word processing has become popular due to its wide range of personal, business, and other applications. ESL learners, like others, need word processing for job search, employment, and personal purposes. Word-processing skills have become the backbone of computer literacy skills. Features PARTS OF THE SCREEN The Word screen can be overwhelming for novice learners. The numerous bars on the screen such as toolbars, scroll bars, and status bar confuse learners who are using Word for the first time. It is important that learners become familiar with parts of the screen and understand the function of each toolbar but we recommend that the Standard and Formatting toolbars as well as the Status bar be hidden for LINC One level. Menu bar Title bar Minimize Restore Button Button Close Word Close current Rulers document Insertion Point (cursor) Vertical scroll bar Editing area Document Status bar Horizontal Views scroll bar A SOFTWARE GUIDE FOR LINC INSTRUCTORS 131 1 MICROSOFT WORD Hiding Standard toolbar, Formatting toolbar, and Status bar: • To hide the Standard toolbar, click View | Toolbars on the Menu bar. Check off Standard. LINC ONE LINC • To hide the Formatting toolbar, click View | Toolbars on the Menu bar. -
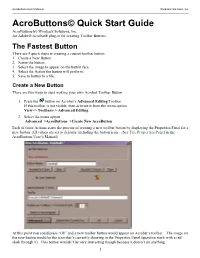
Acrobuttons Quick Start Tutorial
AcroButtons User’s Manual WindJack Solutions, Inc. AcroButtons© Quick Start Guide AcroButtons by Windjack Solutions, Inc. An Adobe® Acrobat® plug-in for creating Toolbar Buttons. The Fastest Button There are 5 quick steps to creating a custom toolbar button.. 1. Create a New Button 2. Name the button. 3. Select the image to appear on the button face. 4. Select the Action the button will perform. 5. Save to button to a file. Create a New Button There are two ways to start making your own Acrobat Toolbar Button. 1. Press the button on Acrobat’s Advanced Editing Toolbar. If this toolbar is not visible, then activate it from the menu option View–> Toolbars–> Advanced Editing. 2. Select the menu option Advanced–>AcroButtons–>Create New AcroButton Each of these Actions starts the process of creating a new toolbar button by displaying the Properties Panel for a new button. All values are set to defaults, including the button icon. (See The Properties Panel in the AcroButtons User’s Manual) At this point you could press “Ok” and a new toolbar button would appear on Acrobat’s toolbar. The image on the new button would be the icon that’s currently showing in the Properties Panel (question mark with a red slash through it). This button wouldn’t be very interesting though because it doesn’t do anything. 1 AcroButtons User’s Manual WindJack Solutions, Inc. Name the Button Change the default name, “Untitled0”, to something more descriptive. For this example change it to “NextPage”. The button’s name is important for several different reasons. -

HTML Viewer Page 5
HTML Report Viewer Version: 18.1 Copyright © 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from, through any means, in parts or in whole, without a prior written permission from Intellicus Technologies. All other product names are believed to be registered trademarks of the respective companies. Dated: August 2015 Acknowledgements Intellicus acknowledges using of third-party libraries to extend support to the functionalities that they provide. For details, visit: http://www.intellicus.com/acknowledgements.htm 2 Contents 1 HTML Report Viewer 4 HTML Viewer page 5 Navigate 6 Table of Contents 7 Refresh the report 8 Collaborate 8 Email 8 Upload 9 Print 10 Publish 12 Export 13 View Alerts 14 3 1 HTML Report Viewer Reports are displayed in HTML Viewer when you select View > HTML option on Report Delivery Options page. Figure 1: View > HTML on Report Delivery Options page In addition to viewing a report, you can carry out following activities on HTML Viewer: • Navigate • Refresh the report • Collaborate • Email • Print • Publish • Export • Upload • View Alerts You can initiate any of these activities by clicking respective tool button from the toolbar. 4 HTML Viewer page Depending on the report being viewed and the steps taken as post-view operations, it may have multiple components on it: Toolbar Explore Pane Page area Figure 2: HTML Viewer HTML Viewer has mainly three components. On the top, it has toolbar having buttons for all the operations that can be done on the Viewer. The Explore pane appears on the left side of the window. -

Introduction to Office 2007 the Ribbon Home
Introduction to Office 2007 Introduction to Office 2007 class will show what’s different in Office 2007. The Ribbon at the top of the page has replaced menus and toolbars in Word, Excel, PowerPoint, Access and new messages in Outlook. This class will cover the use of the Ribbon, the Office Button (where is the File menu?), getting Help and online Training, the Mini Toolbar, the Quick Access Toolbar, new File Formats, and a Few Fun Features. The class and short handout are designed so when you return to your desk you can begin using the Office 2007 products. There are three major differences in Office 2007, the Ribbon, the Office Button and the new file formats. The Ribbon The Ribbon at the top of the page has replaced menus and toolbars in Word, Excel, PowerPoint, Access and new messages in Outlook. Commands – Are buttons, menus or boxes where you enter information. Groups – Are sets of related commands. Tabs – Represent core tasks. Home Tab Try it: 1. Start Word 2007, Excel 2007 & PowerPoint 2007. 2. Click on each tab to display different groups of commands. 3. Hover over a command for Enhanced Toolbar Tips. Note: keyboard shortcuts are shown if available. The Home Tab displays the most commonly used commands. In Word and Excel these include Copy, Cut, and Paste, Bold, Italic, Underscore etc. The commands are arranged in groups: Clipboard, Font, Paragraph, Styles and Editing. The most frequently used commands, Paste, Cut and Copy, are the left most in the first group in the Home Tab. The less frequently used commands or command choices can Try it: 1. -

Remove ANY TOOLBAR from Internet Explorer, Firefox and Chrome
Remove ANY TOOLBAR from Internet Explorer, Firefox and Chrome Browser toolbars have been around for years, however, in the last couple of months they became a huge mess. Unfortunately, lots of free software comes with more or less unwanted add-ons or browser toolbars. These are quite annoying because they may: Change your homepage and your search engine without your permission or awareness Track your browsing activities and searches Display annoying ads and manipulate search results Take up a lot of (vertical) space inside the browser Slow down your browser and degrade your browsing experience Fight against each other and make normal add-on handling difficult or impossible Become difficult or even impossible for the average user to fully uninstall Toolbars are not technically not a virus, but they do exhibit plenty of malicious traits, such as rootkit capabilities to hook deep into the operating system, browser hijacking, and in general just interfering with the user experience. The industry generally refers to it as a “PUP,” or potentially unwanted program. Generally speaking, toolbars are ad-supported (users may see additional banner, search, pop-up, pop-under, interstitial and in-text link advertisements) cross web browser plugin for Internet Explorer, Firefox and Chrome, and distributed through various monetization platforms during installation. Very often users have no idea where did it come from, so it’s not surprising at all that most of them assume that the installed toolbar is a virus. For example, when you install iLivid Media Player, you will also agree to change your browser homepage to search.conduit.com, set your default search engine to Conduit Search, and install the AVG Search-Results Toolbar. -

Geotool Software User Tutorial
IDC/SA/SI February, 2013 English only Geotool Software User Tutorial This document contains the Geotool software user tutorial. The tutorial is divided into a set of exercises that can be followed individually. Together, the exercises cover the full functionality of the system. Summary Geotool is a software system that allows a user to interactively display and process seismoacoustic data from International Monitoring System (IMS) stations. The software can be customised and extended. This tutorial instructs users how to use the basic features of Geotool including data input, basic waveform handling, filters, and arrivals. Geotool has many other capabilities that are not covered here. IDC/SA/SI Page 2 Document history Version Date Author Description 1.0 13.07.2007. Vera Miljanovic Software User Tutorial 2.0 12.08.2009. Vera Miljanovic Updated Tutorial and rewrote Arrivals section 3.0 13.05.2010. Vera Miljanovic Updated Tutorial 4.0 12.10.2010. Vera Miljanovic Updated Tutorial 5.0 10.11.2011 Vera Miljanovic Revisions based on the latest software version 6.0 07.03.2012 Vera Miljanovic Revisions based on the latest software version 7.0 23.01.2013 Vera Miljanovic Revisions based on the latest software version 8.0 08.02.2013 Vera Miljanovic Added Add Station section 9.0 18.02.2013 Remmy Phiri Added Print bulletin section IDC Page 3 Contents SUMMARY 1 DOCUMENT HISTORY 2 CONTENTS 3 1. SCOPE 7 1.1. Identification 7 1.2. System overview 7 1.3. Document overview 7 2. INTRODUCTION 8 2.1. Getting Started with Geotool 8 SECTION 1. -

Microsoft Windows 7
IT Services Training Guide Microsoft Windows 7 IT Services Training Team The University of Manchester email: [email protected] www.itservices.manchester.ac.uk/trainingcourses/coursesforstaff Version 1.0 Table of Contents Starting and Exiting Windows ......................................................................................... 6 Objectives .................................................................................................................... 6 Starting your computer and logging on ......................................................................... 6 Restarting your computer .............................................................................................. 8 Ending your Windows session ...................................................................................... 10 Locking your computer ............................................................................................... 10 Logging off ................................................................................................................. 10 Putting the computer to sleep ..................................................................................... 11 Shutting down your computer ..................................................................................... 11 The Desktop ............................................................................................................... 11 Objectives ................................................................................................................. -
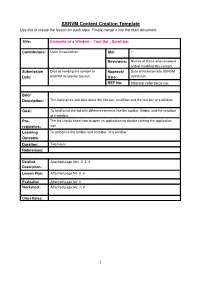
SSRVM Content Creation Template Use This to Create the Lesson for Each Topic
SSRVM Content Creation Template Use this to create the lesson for each topic. Finally merge it into the main document. Title: Elements of a Window : Tool Bar , Scroll bar Contributors: Usha Viswanathan Std: 1 Reviewers: Names of those who reviewed and/or modified this content. Submission Date of sending the content to Approval Date of inclusion into SSRVM Date: SSRVM Academic Council. Date: curriculum. REF No: Internal reference no. Brief Description: This topic gives and idea about the title bar, scroll bar and the tool bar of a window. Goal: To familiarise the kid with different elements like the toolbar, titlebar and the scrollbar of a window. Pre- The kid should know how to open an application by double clicking the application requisites: icon. Learning To recognise the toolbar and scrollbar of a window. Outcome: Duration: Two hours References: Detailed Attached page Nos. 2, 3, 4 Description: Lesson Plan: Attached page No. 5, 6 Evaluation Attached page No. 6 Worksheet: Attached page No. 7, 8 Other Notes: 1 Detailed Description: Tool Bar and Scroll Bars Many programs and applications run within windows that can be opened, minimized, resized and closed (See figure (a)). The top bar of the window is called the Title Bar. This horizontal area at the top of a window identifies the window. The title bar also acts as a handle for dragging the window. figure(a) Tool Bar A toolbar is a set of tools that are grouped together into an area on the main window of an application. Typically, the tools are arranged into a horizontal strip called a bar, hence the term toolbar. -

Adobe® Acrobat® 9 Pro Accessibility Guide: Creating Accessible Forms
Adobe® Acrobat® 9 Pro Accessibility Guide: Creating Accessible Forms Adobe, the Adobe logo, Acrobat, Acrobat Connect, the Adobe PDF logo, Creative Suite, LiveCycle, and Reader are either registered trade- marks or trademarks of Adobe Systems Incorporated in the United States and/or other countries. AutoCAD is either a registered trade- mark or a trademark of Autodesk, Inc., in the USA and/or other countries. GeoTrust is a registered trademark of GeoTrust, Inc. Microsoft and Windows are either registered trademarks or trademarks of Microsoft Corporation in the United States and/or other countries. All other trademarks are the property of their respective owners. © 2008 Adobe Systems Incorporated. All rights reserved. Contents Contents i Acrobat 9 Pro Accessible Forms and Interactive Documents 3 Introduction 3 PDF Form Fields 3 Use Acrobat to Detect and Create Interactive Form Fields 3 Acrobat Form Wizard 4 Enter Forms Editing Mode Directly 4 Create Form Fields Manually 5 Forms Editing Mode 5 Creating a New Form Field 6 Form Field Properties 7 Tooltips for Form Fields 7 Tooltips for Radio Buttons 8 Editing or Modifying an Existing Form Field 9 Deleting a Form Field 9 Buttons 10 Set the Tab Order 10 Add Other Accessibility Features 11 ii | Contents Making PDF Accessible with Adobe Acrobat 9 Pro Making PDF Accessible with Adobe Acrobat 9 Pro Acrobat 9 Pro Accessible Forms and Interactive Documents Introduction Determining if a PDF file is meant to be an interactive form is a matter of visually examining the file and looking for the presence of form fields, or areas in the document where some kind of information is being asked for such as name, address, social security number. -

Using a Scroll-Wheel Pen in Complex Virtual Environment Applications
Wheelie - Using a Scroll-Wheel Pen in Complex Virtual Environment Applications M. Wögerbauer A. L. Fuhrmann VRVis – Research Center for Virtual Reality and VRVis – Research Center for Virtual Reality and Visualization, Ltd Visualization, Ltd Donau-City-Strasse 1 Donau-City-Strasse 1 A-1220, Vienna, Austria A-1220, Vienna, Austria [email protected] [email protected] ABSTRACT Input devices and system control techniques for complex virtual environment (VE) applications are still an open field of research. We propose the use of a scroll-wheel as an extra, dedicated input stream on a tracked stylus for means of system control. We demonstrate how this enhanced stylus can be used together with an appropriate user interface to quickly select commands, change tools, and adjust parameters. This user interface consists of two different styles: a toolbar and a graphical menu system, both accessible by the same hand that holds the stylus. The scroll-wheel extension does in no way impair the conventional use of the stylus. Keywords Input Devices, Scrolling, Virtual environments, Application control, System control, Tool selection, Menu system, Responsive Workbench, 3D Interaction 1. INTRODUCTION AND Navigation changes the viewpoint in the environ- MOTIVATION ment, and can further be divided into a cognitive part Virtual environments (VEs) promise a great amount (wayfinding) and a motor part (travel). Selection re- of potential as a working environment for applica- fers to the task of choosing one or more objects from tions with a high degree of interactive complexity. a set which is closely related to the third task of ma- They provide a virtual workspace for the user in nipulation.