Evaluation of Mid-Gut Bacteria Present in the Larva of Rice Moth
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Efficacy of Azadirachta Indica and Eucalyptus Globulus Against Corcyra Cephalonica
Int. J. Adv. Res. Biol. Sci. (2019). 6(7): 119-123 International Journal of Advanced Research in Biological Sciences ISSN: 2348-8069 www.ijarbs.com DOI: 10.22192/ijarbs Coden: IJARQG(USA) Volume 6, Issue 7 -2019 Research Article DOI: http://dx.doi.org/10.22192/ijarbs.2019.06.07.015 Efficacy of Azadirachta indica and Eucalyptus globulus against Corcyra cephalonica Harminder Singh, Anita Singh* and Jora Singh Brar Department of Entomology, University College of Agriculture, Guru Kashi University, Talwandi Sabo, Bathinda Punjab-151302 Email ID*: [email protected] Abstract The four different concentrations (0.5, 1.0, 1.5 and 2.0 g) of Azadirachta indica and Eucalyptus globules powdered leaves were tested against all stages of rice-moth larvae. The result revealed that maximum mortality were observed at higher concentration (2.0 g) with both botanicals in first and second instar of Corcyra cephalonica. When compared both botanicals, E. globulus shows more effectiveness as compared to Azadirachta indica on larvae of rice moth. Therefore on small scale these plant powders having the insecticidal effect could be used as good alternatives for synthetic pesticides against this pest. Keywords: Corcyra cephalonica, Eucalyptus globulus, Azadirachta indica, Mortality Introduction C. cephalonica causes serious damage to stored grain by feeding, leaving dense and tough silken threads Corcyra cephalonica (Stainton) (Lepidoptera: (Ayyar, 1934; Prevett, 1964). The damage also form Pyralidae) is one of the most widely distributed critical frass which make grains useless for human insect pests of the stored grains (Osmen, 1984). It is consumption (Frenemore and Prakash, 1992). To commonly known as Rice moth. -

Stored Grain Insects and Pea Weevil (Live) Insects Large – Dead Or Alive
To whom it may concern, Proposal for GTA Standards change regarding Cereal grains for categories: Stored Grain Insects and Pea Weevil (live) Insects Large – dead or alive Currently there is a lack of reference with insects of NIL tolerance applied by DA for export and that listed within GTA standards. This has the potential to cause contract disputes especially in the grower direct to port transactions. At present if a supplier delivers grain with live insects for example Small-eyed flour beetles and Black fungus beetles, there is no reference in the standards that declare such insects as NIL tolerance. If the buyer was loading a container direct for export this would pose a problem due to the NIL tolerance being applied by DA for export phytosanitary requirements. These insects are in the same category as Psocids which are listed in GTA receival standards. I would like to see the GTA "Stored Grain Insects and Pea Weevil (live)" & "Insects Large – dead or alive" reflect the Department of Agriculture PEOM 6a: Pests, Diseases and Contaminants of Grain and Plant Products (excluding horticulture) http://www.agriculture.gov.au/SiteCollectionDocuments/aqis/exporting/plants-exports-operation-manual/vol6A.pdf I put forward the motion to have all major and minor injurious pests listed within PEOM 6a that apply to cereal grains to be of NIL tolerance within the GTA standards. 1) This would involve moving the Hairy Fungus Beetle Typhaea stercorea from “Insects Large – dead or alive” to the list of “Stored Grain Insects and Pea Weevil (live)”. Thus taking it from a tolerance level of 3 per half litre to NIL. -

Effect of Various Cereals on the Development of Corcyra Cephalonica (Stainton ) and Its Egg Parasitoid Trichogramma Chilonis (Ishii) M
ISSN 0258-7122 (Print), 2408-8293 (Online) Bangladesh J. Agril. Res. 41(1): 183-194, March 2016 EFFECT OF VARIOUS CEREALS ON THE DEVELOPMENT OF CORCYRA CEPHALONICA (STAINTON ) AND ITS EGG PARASITOID TRICHOGRAMMA CHILONIS (ISHII) M. NASRIN1, M. Z. ALAM2, S. N. ALAM3 M. R. U. MIAH4 AND M. M. HOSSAIN5 Abstract Eight types of cereals viz., wheat grain, chopped wheat, paddy grain, rice grain, maize grain, chopped maize, rice bran, mixture of rice bran and chopped rice were fed to observe the development parameters like egg, larva, pupa and adult stages of Corcyra cephalonica (stainton) for three consecutive generations. The parasitism efficiency of Trichogramma chilonis (Ishii) was also evaluated on the resultant host eggs of C. cephalonica. The C. cephalonica revealed the highest number of eggs (115.6 female1), higher hatchability (92.9%), extented larval duration (45.9 days), increased larval weight (0.058 gm), survival rate (88.3%), adult emergence rate (93.5%), and male and female longevity (7.7, 7.2 days respectively) when they were reared on chopped wheat. On the other hand, the lowest number of egg was found on paddy husk (29.2 female-1). The lowest hatchability (45.6%), larval duration (45.9 days), larval weight (0.029gm), and survival rate (38.2%), pupal duration (17.9 days) adult emergence (42.0%), male and female longevity (4.8 and 4.7 days respectively) were found on paddy husk. The effect of food materials also reflected on the parasitism efficiency of the egg parasitoid T. chilonis. The highest percent egg parasitization was done by the T. -

Biodiversity and Faunistic Studies of the Family Pyralidae
Pak. j. life soc. Sci. (2017), 15(2): 126-132 E-ISSN: 2221-7630;P-ISSN: 1727-4915 Pakistan Journal of Life and Social Sciences www.pjlss.edu.pk RESEARCH ARTICLE Biodiversity and Faunistic Studies of the Family Pyralidae (Lepidoptera) from Pothwar Region, Punjab, Pakistan Tazeel Ahmad 1, Zahid Mahmood Sarwar 2*, Mamoona Ijaz 2 , Muhammad Sajjad and Muhammad Bin yameen 2 1 Pir Mehr Ali Shah -Arid Agriculture University Rawalpindi 2 Department of Entomology, Bahauddin Zakariya University, Multan 60800, Pakistan ARTICLE INFO ABSTRACT Received: Jan 24, 2017 Snout moths (Lepidoptera: Pyralidae) are economic pests of agricultural crops and Accepted: Aug 18, 2017 forest plantations. To explore new species via taxonomic identification, 127 specimens of moths were collected from different areas of Pothwar region of Keywords Pakistan including Chakwal, Attock, Jhelum and Rawalpindi districts during 2009- Pothwar 2010. The characters of the specimens were identified at species level by using Pyralidae Hampson’s key and other taxonomic resources. All of these species new to Pyralid Snout Moths moth fauna of Pothwar region namely: Galleria mellonella Linnaeus., Achroia Taxonomic keys grisella Fabricius., Corcyra cephalonica Staint., Chilo simplex Butler., Scirpophaga auriflua Zeller., S. chrysorrhoa Zeller., Leucinodes orbonalis Guenee., Zinckenia fascialis Cramer and Cnaphalocrocis medinalis Guenee. are reported for the first time from Pothwar region. To understand the biodiversity of moths, distribution of the species were also studied for each district of Pothwar region. The distribution pattern of Corcyra cephalonica (42), Leucinodes orbonalis (18), Scirpophaga auriflua (10) and Scirpophaga chrysorrhoa (9), Chilo simplex (6), Zinckenia fascialis (7), Cnaphalocrocis medinalis (10), Galleria mellonella (24) and Achroia *Corresponding Author: grisella (1) was observed. -

Toxicity, Antifeedant, Egg Hatchability and Adult Emergence Effect of Piper Nigrum L. and Jatropha Curcas L. Extracts Against Ri
Vol. 7(18), pp. 1255-1262, 10 May, 2013 DOI: 10.5897/JMPR12.600 Journal of Medicinal Plants Research ISSN 1996-0875 ©2013 Academic Journals http://www.academicjournals.org/JMPR Full Length Research Paper Toxicity, antifeedant, egg hatchability and adult emergence effect of Piper nigrum L. and Jatropha curcas L. extracts against rice moth, Corcyra cephalonica (Stainton) Mousa Khani 1,2 , Rita Muhamad Awang 2*, Dzolkhifli Omar 2 and Mawardi Rahmani 3 1Cultivation and Development Department of Medicinal Plants Research Center, Institute of Medicinal Plants, ACECR, Karaj, Iran. 2Department of Plant Protection, Faculty of Agriculture, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia. 3Department of Chemistry, Faculty of Science, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia. Accepted 8 August, 2012 Petroleum ether extract of black pepper, Piper nigrum and physic nut, Jatropha curcas were shown to have insecticidal efficacies against rice moth, Corcyra cephalonica (Stainton). The C. cephalonica 3rd instar larvae were shown to have similarities susceptibility to petroleum ether extract of Piper nigrum and J. curcas with LC 50 values of 12.52 and 13.22 µL/ml, respectively. In a bioassay using no-choice tests, the parameters used to evaluate antifeedant activity were relative growth rate (RGR), relative consumption rate (RCR), efficiency on conversion of ingested food (ECI) and grain protection or feeding deterrence indices (FDI). Both extracts showed high bioactivity at all doses against C. cephalonica larvae and antifeedant action was increased with increasing plant extract concentrations. The petroleum ether extract of P. nigrum and J. curcas showed strong inhibition on egg hatchabilities and adult emergence of C. -

View Full Text-PDF
Int.J.Curr.Res.Aca.Rev.2015; 3(4):373-380 International Journal of Current Research and Academic Review ISSN: 2347-3215 Volume 3 Number 4 ( April-2015) pp. 373-380 www.ijcrar.com Variation in biological parameters of Corcyra cephalonica Stainton (Lepidoptera:Pyralidae) with quality of rearing media N. Chaudhuri1*, A. Ghosh2 and J. Ghosh3 1Department of Agricultural Entomology, RRS, Terai Zone, Uttar Banga Krishi Viswavidyalaya, Pundibari, Cooch Behar-736165, West Bengal, India 2Department of Agricultural Statistics, RRS, Terai Zone, Uttar Banga Krishi Viswavidyalaya, Pundibari, Cooch Behar-736165, West Bengal, India 3Department of Agricultural Entomology, Uttar Banga Krishi Viswavidyalaya, Pundibari, Cooch Behar-736165, West Bengal, India *Corresponding author KEYWORDS ABSTRACT Food grains, Variation in biological parameters of Corcyra cephalonica Stainton dextrose, (Lepidoptera:Pyralidae) with quality rearing media was studied against four yeast, (4) different types of locally available grains namely maize, wheat, Italian larva, millet and scented rice as solo and fortified with 3% dextrose and yeast. The pupa, insect had significantly shortest larval-pupal period when the food grains adult, were fortified with dextrose followed by yeast and the period was longest in Corcyra solo grains. The insect complete its life cycle within 51.28 days during cephalonica August-October as compared to 54.67 days in May-August. The period was longest in scented rice (64.71 days) and shortest in maize-dextrose (45.00 days). Adult emergence was the maximum during August-October (71.33%) and in wheat (solo as well as fortified). The sex ratio showed female dominance. Fortification of 3% dextrose and yeast increased female per cent and thereby fecundity in Italian millet. -
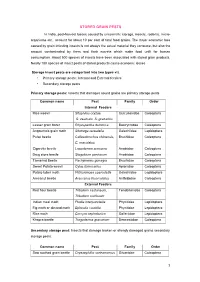
Stored Grain Pests
STORED GRAIN PESTS In India, post-harvest losses caused by unscientific storage, insects, rodents, micro- organisms etc., account for about 10 per cent of total food grains. The major economic loss caused by grain infesting insects is not always the actual material they consume, but also the amount contaminated by them and their excreta which make food unfit for human consumption. About 500 species of insects have been associated with stored grain products. Nearly 100 species of insect pests of stored products cause economic losses Storage insect pests are categorized into two types viz. • Primary storage pests : Internal and External feeders • Secondary storage pests Primary storage pests: Insects that damages sound grains are primary storage pests Common name Pest Family Order Internal Feeders Rice weevil Sitophilus oryzae, Curculionidae Coleoptera S. zeamais, S. granarius Lesser grain borer Rhyzopertha dominica Bostrychidae Coleoptera Angoumois grain moth Sitotroga cerealella Gelechiidae Lepidoptera Pulse beetle Callosobruchus chinensis, Bruchidae Coleoptera C. maculatus Cigarette beetle Lasioderma sericorne Anobiidae Coleoptera Drug store beetle Stegobium paniceum Anobiidae Coleoptera Tamarind Beetle Pachymeres gonagra Bruchidae Coleoptera Sweet Potato weevil Cylas formicarius Apionidae Coleoptera Potato tuber moth Phthorimoea operculella Gelechiidae Lepidoptera Arecanut beetle Araecerus fasciculatus Anthribidae Coleoptera External Feeders Red flour beetle Tribolium castaneum, Tenebrionidae Coleoptera Tribolium confusum Indian meal -

A Theoretical Study on the Sex-Pheromones of the Rice Moth, Corcyra Cephalonica Stainton
Tr. J. of Biology 22 (1998) 229-232 © TÜBİTAK A Theoretical study on the Sex-pheromones of the Rice Moth, Corcyra cephalonica Stainton Lemi TÜRKER Middle East Technical University Department of Chemistry Ankara-TURKEY Recived: 08.05.1997 Abstract: Molecular orbital calculations (AM1 type) were performed on the geometry optimized structures of various farnesal isomers and methyl esters of farnesoic acids. It has been found that the major component, E, E-farnesal, of the sex-pheromone for the rice moth, Corcyra cephalonica Stainton is chemically the most exothermic and stable of all the farnesal isomers. Then, the resem- blance of molecular shapes was detrmined in order to design more stable sex-attractants. Key Words: Corcyra cephalonica, Pyralidae, Galleriinae, molecular orbital calculations, pheromones. Prinç Güvesi Corcyra cephalonica Stainton’un Seks Feromonları Üzerine Teorik bir Araştırma Özet: Geometry optimize edilmiş çeşitli farnesal izomerleri ve farnesoic acid metil esterleri üzerinde moleküler yörünge hesaplamaları (AM1 tipi) yapılmıştır. Pirinç güvesi, Corcyra cephalonica. Stainton’un seks-feromonunun esas elemanı olan E, E-farnesal’ın, bütün farnesal izomerleri içinde kimyasal olarak en ekzotermik ve dayanaklı olduğu bulunmuştur. Müteakiben, daha dayanıklı seks-çekicileri tasarımı amacı ile, molekülsel şekil benzerlikleri tayin edilmiştir. Anahtar Sözcükler: Corcyra cephalonica, Pyralidae. Galleriinae, moleküler yörünge hesapları, fer- omonlar. Introduction The rice moth, Corcyra cephalonica Stainton (Pyralidae, Galleriinae) is a major pest of stored products and quite often its population thus its harm reaches very unplesant level in subtropical countries like India that, some extensive research has been carried out about the biology of this insect as well as different aspects of the sex pheromone (1, 2). -

Development of an Optimum Diet for Mass Rearing of The
Journal of Insect Science, (2019) 19(2): 1; 1–5 doi: 10.1093/jisesa/iez020 Research Development of an Optimum Diet for Mass Rearing of the Downloaded from https://academic.oup.com/jinsectscience/article-abstract/19/2/1/5366655 by University Libraries | Virginia Tech user on 02 April 2019 Rice Moth, Corcyra cephalonica (Lepidoptera: Pyralidae), and Production of the Parasitoid, Habrobracon hebetor (Hymenoptera: Braconidae), for the Control of Pearl Millet Head Miner Laouali Amadou,1,2 Ibrahim Baoua,2 Malick N. Ba,3 and Rangaswamy Muniappan4,5 1Institut National de la Recherche Agronomiques du Niger (INRAN), CERRA de Maradi, BP 240 Maradi, Niger, 2Université Dan Dicko Dankoulodo de Maradi, BP 465 Maradi, Niger, 3International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Niamey, Niger BP 12404, Niamey, Niger, 4IPM Innovation Lab, Virginia Tech, 526 Prices Fork Road, Blacksburg, VA 24061-0378, and 5Corresponding author, e-mail: [email protected] Subject Editor: Muhammad Chaudhury Received 17 October 2018; Editorial decision 1 February 2019 Abstract The rice moth, Corcyra cephalonica Stainton, an alternate host for the production of the parasitoid, Habrobracon hebetor Say, was reared on different diets, including pearl millet [Pennisetum glaucum (L.) R. Br.] (Poales: Poaceae) flour only, and in combinations of flours of sorghumSorghum [ bicolor (L.) Moench] (Poales: Poaceae), peanut (Arachis hypogea L.) (Fabales: Fabaceae), and cowpea [Vigna unguiculata (L.) Walp.] (Fabales: Fabaceae) to identify the optimal and economical proportion to be used under the conditions of Niger. The addition of cowpea or peanut to the pearl millet diet slightly increased C. cephalonica larval development time. Likewise, the addition of cowpea or peanut to cereal diets yielded a higher C. -

Novel Attractants of Galleria Mellonella L (Lepidoptera Pyralidae Galleriinae) L Türker, I Togan, S Ergezen, M Özer
Novel attractants of Galleria mellonella L (Lepidoptera Pyralidae Galleriinae) L Türker, I Togan, S Ergezen, M Özer To cite this version: L Türker, I Togan, S Ergezen, M Özer. Novel attractants of Galleria mellonella L (Lepidoptera Pyralidae Galleriinae). Apidologie, Springer Verlag, 1993, 24 (4), pp.425-430. hal-00891087 HAL Id: hal-00891087 https://hal.archives-ouvertes.fr/hal-00891087 Submitted on 1 Jan 1993 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Original article Novel attractants of Galleria mellonella L (Lepidoptera Pyralidae Galleriinae) L Türker I2 Togan S Ergezen3 M Özer1 1 Middle East Technical University, Department of Chemistry, Ankara; 2 Middle East Technical University, Department of Biology, Ankara; 3 Marmara Universitesi, Egitim Fakültesi Bioloji Bölümü, Istanbul, Turkey (Received 20 January 1993; accepted 14 April 1993) Summary — Various synthetic esters were tested for their attractiveness to females of Galleria mel- lonella in comparison with n-nonanal, a component of the sex pheromone of Galleria mellonella. It has been shown that methyl octanoate, ethyl octanoate and methyl decanoate attract females. Parti- cularly, methyl octanoate is as effective as n-nonanal in bioassay and could be a potential pest control agent. -

Assessment of Damage by the Rice Moth, Corcyra Cephalonica (81.) on Different Grains at Four Levels of Moisture Content
Pertanika 7(3), 53-58 (1984) Assessment of Damage by the Rice Moth, Corcyra cephalonica (81.) on Different Grains at Four Levels of Moisture Content. NOORMA OSMAN Department ofPlant Protection, Faculty ofAgriculture Universiti Pertanian Malaysia, Serdang, Selangor, Malaysia. Key words: Gorcyra cephalonica; damage assessment RINGKASAN Taksiran kerosakan oleh C.cephalonica ke atas 'millet', sekoi, beras dan padi dalam keadaan peratus kandungan kelembapan yang berlainan telah ditentukan dengan menggunakan kaedah 'count and weigh method'. Parameter-parameter yang ditentukan adalah peratus kerosakan pada bijirin, peratus kehilangan berat kering serta berat kering '!rass' dan 'webbings'. Peratus larva yang berumur 12 ± 12 jam yang dapat meneruskan he peringkat dewasa dan purata kala perkembangan larva ke peringkat dewasa juga ditentukan. Empat peringkat peratus kandungan kelembapan yang digunakan dalam kajian ini tidak mempengaruhi peratus kerosakan pada bijirin, peratus kehilangan oerat kering serta berat kering 'fras' dan 'webbings'. Padi ialah yang terkurang sekali kerosakannya; sedikit sekali larva yang dapat meneruskan ke peringkat dewasa. Peratus larva yang dapat meneruskan ke peringkat dewasa, meningkat daripada peratus kan.dungan kelembapan 11.1% ke 13.2% bagi 'millet', sekoi dan beras, sementarapadaperatuskandungan kelembapan 14.1%, ia susut. Peratus larva yang dapat meneruskan ke peringkat dewasa pada beras yang kandungan kelembapan 11.0% berbeza dengan jelas, dibandingkan dengan beras yang kandungan kelembapannya 14.1%. Puratakala perkembangan larva pada 'millet', sekoi dan beras yang kandungan kelembapan 11.1% berbeza dengan jelas dibandingkan dengan kandungan kelembapan 14.0%. SUMMARY Assessment ofdamage by C.cephalonica on millet, sorghum, milled rice and padi at four moisture contents was determined using the count and weigh method. The parameters determined were percent damaged kernels, percent dry weight loss and dry weight of frass and webbings. -

Impact of Abiotic Factors on the Growth and Development of Corcyra Cephalonica Stainton in Stored Maize
Int.J.Curr.Microbiol.App.Sci (2017) 6(5): 1599-1608 International Journal of Current Microbiology and Applied Sciences ISSN: 2319-7706 Volume 6 Number 6 (2017) pp. 1599-1608 Journal homepage: http://www.ijcmas.com Original Research Article https://doi.org/10.20546/ijcmas.2017.606.188 Impact of Abiotic Factors on the Growth and Development of Corcyra cephalonica Stainton in Stored Maize H.R. Meena, A. Meena, A. Kumar, A.K. Meena*, S.K. Chauhan and B.M. Meena Department of Entomology, Rajasthan College of Agriculture Maharana Pratap University of Agriculture and Technology, Udaipur- 313001, Rajasthan, India *Corresponding author ABSTRACT An experiment was conducted in the laboratory to find out the impact of temperature and relative humidity on growth and development of C. cephalonica during, 2010-2011, revealed that the development period of male and female was maximum with a mean of K e yw or ds 89.50 and 92.00 days at 20°C temperature and 40 per cent relative humidity, respectively. Abiotic, Corcyra The maximum larval period of (70.10 days) was recorded at 20°C temperature and 40 per cephalonica, cent relative humidity; whereas, the maximum weight of full grown larva (51.00 mg) was Temperature , recorded at 30°C temperature and 80 per cent relative humidity. The maximum pupal Relative humidity, period of male (13.60 days) and female (14.40 days) were observed at 20°C temperature Larvae. and 40 per cent relative humidity, while the higher weight of male (36.00 mg) and female Article Info (38.00 mg) pupa were observed at 30°C temperature and 80 per cent relative humidity.