Positive AR Female
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T
Mutation of the Fumarase Gene in Two Siblings with Progressive Encephalopathy and Fumarase Deficiency T. Bourgeron,* D. Chretien,* J. Poggi-Bach, S. Doonan,' D. Rabier,* P. Letouze,I A. Munnich,* A. R6tig,* P. Landneu,* and P. Rustin* *Unite de Recherches sur les Handicaps Genetiques de l'Enfant, INSERM U393, Departement de Pediatrie et Departement de Biochimie, H6pital des Enfants-Malades, 149, rue de Sevres, 75743 Paris Cedex 15, France; tDepartement de Pediatrie, Service de Neurologie et Laboratoire de Biochimie, Hopital du Kremlin-Bicetre, France; IFaculty ofScience, University ofEast-London, UK; and IService de Pediatrie, Hopital de Dreux, France Abstract chondrial enzyme (7). Human tissue fumarase is almost We report an inborn error of the tricarboxylic acid cycle, fu- equally distributed between the mitochondria, where the en- marase deficiency, in two siblings born to first cousin parents. zyme catalyzes the reversible hydration of fumarate to malate They presented with progressive encephalopathy, dystonia, as a part ofthe tricarboxylic acid cycle, and the cytosol, where it leucopenia, and neutropenia. Elevation oflactate in the cerebro- is involved in the metabolism of the fumarate released by the spinal fluid and high fumarate excretion in the urine led us to urea cycle. The two isoenzymes have quite homologous struc- investigate the activities of the respiratory chain and of the tures. In rat liver, they differ only by the acetylation of the Krebs cycle, and to finally identify fumarase deficiency in these NH2-terminal amino acid of the cytosolic form (8). In all spe- two children. The deficiency was profound and present in all cies investigated so far, the two isoenzymes have been found to tissues investigated, affecting the cytosolic and the mitochon- be encoded by a single gene (9,10). -

Supplementary Table 1. Biochemical/Metabolic Disorders in ICEE; Clinical Features and Relevant Diagnostic Tests
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Arch Dis Child Educ Pract Ed Supplementary table 1. Biochemical/metabolic disorders in ICEE; clinical features and relevant diagnostic tests: Presentation Age of </> 1 year of Disorder group Disorder Sub disorder onset age Other features Treatable? Test Often infants born prem, sometimes family history of < 3 infertility & recurrent CSF pyridoxal Vitamin B6 PNPO months <1y miscarriage. Yes phosphate (UOA) Metabolic acidosis, electrolyte disturbance, abdominal distension, and feed intolerance, CSF pipecolate (pre- resulting in misdiagnosis as treatment); (urine Pyridoxine responsive < 3 hypoxic–ischaemic AASA where epilepsy months <1y encephalopathy or sepsis. Yes available) Failure to thrive & multisystem disease in early infancy; Congenital disorders of </> 3 hypotonia and seizures can glycosylation. Many subtypes months </> 1y be part of the clinical picture. most not Transferrin glycoforms Methylmalonic </> 3 Acute metabolic Organic acidurias acidurias months </> 1y decompensations. Yes UOA / acylcarnitines </> 3 Acute metabolic Propionic acidaemia months </> 1y decompensations. Yes UOA / acylcarnitines Developmental delay, hypotonia, dysmorphic D-2-hydroxyglutaric </> 3 features, often with aciduria months </> 1y cardiomyopathy. No UOA L-2-hydroxyglutaric > 3 aciduria months </> 1y Cerebellar dysfunction. Yes UOA Severe muscle weakness, respiratory distress Combined L and D < 3 syndrome, lack of 2OH-glutaric aciduria months <1y psychomotor development. No UOA Perry LD, et al. Arch Dis Child Educ Pract Ed 2021;0:1–8. doi: 10.1136/archdischild-2020-320606 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Arch Dis Child Educ Pract Ed Supplementary table 1. -

Table S1. Disease Classification and Disease-Reaction Association
Table S1. Disease classification and disease-reaction association Disorder class Associated reactions cross Disease Ref[Goh check et al. -

Fumarase Deficiency in Dichorionic Diamniotic Twins
Twin Research and Human Genetics Volume 16 Number 6 pp. 1117–1120 C The Authors 2013 doi:10.1017/thg.2013.72 Fumarase Deficiency in Dichorionic Diamniotic Twins Simone Tregoning,1,2 Wendy Salter,1,2 David R. Thorburn,1,2,3 Miranda Durkie,5 Maria Panayi,5 Joyce Y. Wu,4 Aaron Easterbrook,6 and David J Coman7,8 1Murdoch Children’s Research Institute, Melbourne, Victoria, Australia 2VCGS Pathology, Melbourne, Victoria, Australia 3Department of Paediatrics, University of Melbourne, Melbourne, Victoria, Australia 4Mater Pathology, Brisbane, Queensland, Australia 5Sheffield Diagnostic Genetic Service, Sheffield, UK 6Mater Children’s Hospital, Brisbane, Queensland, Australia 7Department of Metabolic Medicine, Royal Children’s Hospital Brisbane, Herston, Queensland, Australia 8Discipline of Paediatrics, The Wesley-St Andrews Clinical School, University of Queensland, Brisbane, Queensland, Australia Fumarase deficiency is a rare autosomal recessive inborn error of metabolism of the Krebs Tricarboxylic Acid cycle. A heavy neurological disease burden is imparted by fumarase deficiency, commonly manifesting as microcephaly, dystonia, global developmental delay, seizures, and lethality in the infantile period. Heterozygous carriers also carry an increased risk of developing hereditary leiomyomatosis and renal cell carcinoma. We describe a non-consanguineous family in whom a dichorionic diamniotic twin pregnancy resulted in twin boys with fumarase deficiency proven at the biochemical, enzymatic, and molecular levels. Their clinical phenotype included hepatic involvement. A novel mutation in the fumarate hydratase gene was identified in this family. Keywords: fumarase deficiency, Krebs tricarboxylic acid cycle, dichorionic diamniotic, leiomyomatosis, renal cell carcinoma, hepatomegaly Fumarate hydratase (FH) deficiency (OMIM 606812) is a atic disease as part of their clinical phenotype, and a novel rare autosomal recessive defect of the Krebs Tricarboxylic mutation was identified in the FH gene. -

Diseases Catalogue
Diseases catalogue AA Disorders of amino acid metabolism OMIM Group of disorders affecting genes that codify proteins involved in the catabolism of amino acids or in the functional maintenance of the different coenzymes. AA Alkaptonuria: homogentisate dioxygenase deficiency 203500 AA Phenylketonuria: phenylalanine hydroxylase (PAH) 261600 AA Defects of tetrahydrobiopterine (BH 4) metabolism: AA 6-Piruvoyl-tetrahydropterin synthase deficiency (PTS) 261640 AA Dihydropteridine reductase deficiency (DHPR) 261630 AA Pterin-carbinolamine dehydratase 126090 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal recessive) 233910 AA GTP cyclohydrolase I deficiency (GCH1) (autosomal dominant): Segawa syndrome 600225 AA Sepiapterin reductase deficiency (SPR) 182125 AA Defects of sulfur amino acid metabolism: AA N(5,10)-methylene-tetrahydrofolate reductase deficiency (MTHFR) 236250 AA Homocystinuria due to cystathionine beta-synthase deficiency (CBS) 236200 AA Methionine adenosyltransferase deficiency 250850 AA Methionine synthase deficiency (MTR, cblG) 250940 AA Methionine synthase reductase deficiency; (MTRR, CblE) 236270 AA Sulfite oxidase deficiency 272300 AA Molybdenum cofactor deficiency: combined deficiency of sulfite oxidase and xanthine oxidase 252150 AA S-adenosylhomocysteine hydrolase deficiency 180960 AA Cystathioninuria 219500 AA Hyperhomocysteinemia 603174 AA Defects of gamma-glutathione cycle: glutathione synthetase deficiency (5-oxo-prolinuria) 266130 AA Defects of histidine metabolism: Histidinemia 235800 AA Defects of lysine and -
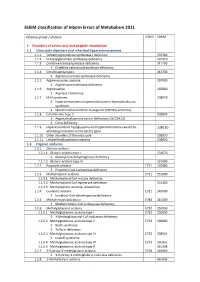
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

1 a Clinical Approach to Inherited Metabolic Diseases
1 A Clinical Approach to Inherited Metabolic Diseases Jean-Marie Saudubray, Isabelle Desguerre, Frédéric Sedel, Christiane Charpentier Introduction – 5 1.1 Classification of Inborn Errors of Metabolism – 5 1.1.1 Pathophysiology – 5 1.1.2 Clinical Presentation – 6 1.2 Acute Symptoms in the Neonatal Period and Early Infancy (<1 Year) – 6 1.2.1 Clinical Presentation – 6 1.2.2 Metabolic Derangements and Diagnostic Tests – 10 1.3 Later Onset Acute and Recurrent Attacks (Late Infancy and Beyond) – 11 1.3.1 Clinical Presentation – 11 1.3.2 Metabolic Derangements and Diagnostic Tests – 19 1.4 Chronic and Progressive General Symptoms/Signs – 24 1.4.1 Gastrointestinal Symptoms – 24 1.4.2 Muscle Symptoms – 26 1.4.3 Neurological Symptoms – 26 1.4.4 Specific Associated Neurological Abnormalities – 33 1.5 Specific Organ Symptoms – 39 1.5.1 Cardiology – 39 1.5.2 Dermatology – 39 1.5.3 Dysmorphism – 41 1.5.4 Endocrinology – 41 1.5.5 Gastroenterology – 42 1.5.6 Hematology – 42 1.5.7 Hepatology – 43 1.5.8 Immune System – 44 1.5.9 Myology – 44 1.5.10 Nephrology – 45 1.5.11 Neurology – 45 1.5.12 Ophthalmology – 45 1.5.13 Osteology – 46 1.5.14 Pneumology – 46 1.5.15 Psychiatry – 47 1.5.16 Rheumatology – 47 1.5.17 Stomatology – 47 1.5.18 Vascular Symptoms – 47 References – 47 5 1 1.1 · Classification of Inborn Errors of Metabolism 1.1 Classification of Inborn Errors Introduction of Metabolism Inborn errors of metabolism (IEM) are individually rare, but collectively numerous. -

Genomeposter2009.Pdf
Fold HumanSelected Genome Genes, Traits, and Landmarks Disorders www.ornl.gov/hgmis/posters/chromosome genomics.energy. -

Beacon Carrier Screening Detecting 400+ Conditions
Beacon Carrier Screening Detecting 400+ Conditions COVERAGE ~99% at 20x DELETION / DUPLICATION ≥ 2 exons TURNAROUND TIME 2 weeks* ACOG/ Expanded Focus Expanded Condition Gene Inheritance ACMG Plus (30 genes†) (401 genes†) (6 genes†) (410 genes†) Alpha thalassemia HBA1/HBA2 AR ü ü ü ü Cystic fibrosis CFTR AR ü ü ü ü Fragile X syndrome FMR1 XL ü ü ü ü Sickle cell disease; Beta thalassemia HBB AR ü ü ü ü Spinal muscular atrophy SMN1 AR ü ü ü ü Bloom syndrome BLM AR - ü ü ü Canavan disease ASPA AR - ü ü ü Congenital disorder of glycosylation type 1a PMM2 AR - ü ü ü ELP1 Familial dysautonomia AR - ü ü ü (IKBKAP) Fanconi anemia group C FANCC AR - ü ü ü Galactosemia GALT AR - ü ü ü Gaucher disease GBA AR - ü ü ü Glycogen storage disease, type 1a G6PC AR - ü ü ü Niemann-Pick disease, type A/B SMPD1 AR - ü ü ü Polycystic kidney disease, PKHD1-related PKHD1 AR - ü ü ü Smith-Lemli-Opitz syndrome DHCR7 AR - ü ü ü Tay-Sachs disease HEXA AR - ü ü ü Tyrosinemia, type 1 FAH AR - ü ü ü Citrullinemia ASS1 AR - ü ü ü Duchenne muscular dystrophy DMD XL - ü ü ü Isovaleric acidemia IVD AR - ü ü ü Medium-chain acyl-CoA dehydrogenase ACADM AR ü ü ü (MCAD) deficiency - Methylmalonic aciduria and homocystinuria, MMACHC AR ü ü ü cblC type - Mucopolysaccharidosis, type I (Hurler syndrome) IDUA AR - ü ü ü Neuronal ceroid lipofuscinosis, CLN3-related CLN3 AR - ü ü ü Phenylalanine hydroxylase deficiency PAH AR ü ü ü (Phenylketonuria) - Pompe disease GAA AR - ü ü ü Rhizomelic chondrodysplasia punctata, type 1 PEX7 AR - ü ü ü Zellweger syndrome, PEX1-related PEX1 AR - ü ü ü * Turnaround time may vary if specimen requires additional confirmatory testing. -

Gene and Disease List
CarrierScreeningGeneandDiseaseList Gene Disease Inheritance Gene Disease Inheritance GRACILEsyndrome;Bjornstadsyndrome;Leigh AAAS Achalasia-addisonianism-alacrimasyndrome AR BCS1L syndrome;MitochondrialcomplexIIIdeficiency, AR nucleartype1 Progressivefamilialintrahepaticcholestasis,type ABCB11 AR BLM Bloomsyndrome AR II ABCC6 Pseudoxanthomaelasticum AR BSND Barttersyndrome,typeIV AR ABCC8 Familialhyperinsulinemichypoglycemiatype1 AR BTD Biotinidasedeficiency AR Desbuquoisdysplasia,typeI;Epiphyseal ABCD1 Adrenoleukodystrophy(X-linked) XL CANT1 AR dysplasia,multiple,7 ACADM MediumchainAcyl-CoAdehydrogenasedeficiency AR CAPN3 Limb-girdlemusculardystrophy,type2A AR ACADS ShortchainAcyl-CoAdehydrogenasedeficiency AR CBS Homocystinuria,CBS-related AR Ushersyndrome,typeID;Deafness,autosomal ACADSB 2-Methylbutyryl-CoAdehydrogenasedeficiency AR CDH23 AR recessive12 Lebercongenitalamaurosis10;Joubertsyndrome ACADVL VerylongchainAcyl-CoAdehydrogenasedeficiency AR CEP290 5; AR Meckelsyndrome4;Senior-Lokensyndrome6 Beta-ketothiolasedeficiency(Alpha- ACAT1 AR CERKL Retinitispigmentosa26 AR methylacetoaceticaciduria) Cysticfibrosis;Congenitalbilateralabsenceofvas ACOX1 Peroxisomalacyl-CoAoxidasedeficiency AR CFTR AR deferens SeverecombinedimmunodeficiencyduetoADA ADA AR CHAT Congenitalmyasthenicsyndrome6 AR deficiency ADAMTS2 EhlersDanlossyndrome,typeVIIC AR CHM Choroideremia AR Congenitalmyasthenicsyndrome4A;Congenital ADAR Aicardi-Goutieressyndrome6 AR CHRNE myasthenicsyndrome4B;Congenitalmyasthenic AR syndrome4C ADGRG1 Bilateralfrontoparietalpolymicrogyria -

WES Gene Package Metabolic Disorders.Xlsx
Whole Exome Sequencing Gene package Metabolic disorders, version 5, 30‐9‐2019 Technical information DNA was enriched using Agilent SureSelect Clinical Research Exome V2 capture and paired‐end sequenced on the Illumina platform (outsourced). The aim is to obtain 8.1 Giga base pairs per exome with a mapped fraction of 0.99. The average coverage of the exome is ~50x. Duplicate reads are excluded. Data are demultiplexed with bcl2fastq Conversion Software from Illumina. Reads are mapped to the genome using the BWA‐MEM algorithm (reference: http://bio‐bwa.sourceforge.net/). Variant detection is performed by the Genome Analysis Toolkit HaplotypeCaller (reference: http://www.broadinstitute.org/gatk/). The detected variants are filtered and annotated with Cartagenia software and classified with Alamut Visual. It is not excluded that pathogenic mutations are being missed using this technology. At this moment, there is not enough information about the sensitivity of this technique with respect to the detection of deletions and duplications of more than 5 nucleotides and of somatic mosaic mutations (all types of sequence changes). HGNC approved Phenotype description including OMIM phenotype ID(s) OMIM median depth % covered % covered % covered gene symbol gene ID >10x >20x >30x AASS Hyperlysinemia, 238700 605113 63 100 97 90 Saccharopinuria, 268700 ABAT GABA‐transaminase deficiency, 613163 137150 100 100 100 96 ABCA1 HDL deficiency, familial, 1, 604091 600046 92 100 100 97 Tangier disease, 205400 ABCB11 Cholestasis, benign recurrent intrahepatic, -

Clinico-Radiological Phenotyping and Diagnostic Pathways in Childhood Neurometabolic Disorders—A Practical Introductory Guide
1230 Review Article on Pediatric Neuroradiology for Trainees and Fellows: An Updated Practical Guide Clinico-radiological phenotyping and diagnostic pathways in childhood neurometabolic disorders—a practical introductory guide Asthik Biswas1,2, Mukul Malhotra3, Kshitij Mankad4, Olivia Carney4, Felice D’Arco4, Karthik Muthusamy3, Sniya Valsa Sudhakar4 1Department of Diagnostic Imaging, The Hospital for Sick Children, Toronto, Ontario, Canada; 2Department of Medical Imaging, University of Toronto, Toronto, Ontario, Canada; 3Department of Neurology, Christian Medical College, Vellore, India; 4Neuroradiology Unit, Great Ormond Street Hospital NHS Foundation Trust, London, UK Contributions: (I) Conception and design: A Biswas, K Muthusamy, SV Sudhakar; (II) Administrative support: K Muthusamy, SV Sudhakar; (III) Provision of study materials or patients: A Biswas, K Muthusamy, SV Sudhakar; (IV) Collection and assembly of data: A Biswas, M Malhotra, K Muthusamy, SV Sudhakar; (V) Data analysis and interpretation: A Biswas, M Malhotra, K Muthusamy, SV Sudhakar; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. Correspondence to: Sniya Valsa Sudhakar. Neuroradiology Unit, Great Ormond Street Hospital NHS Foundation Trust, London, UK. Email: [email protected]. Abstract: Inborn errors of metabolism (IEM) although individually rare, together constitute a significant proportion of childhood neurological disorders. Majority of these disorders occur due to deficiency of an enzyme in a specific metabolic pathway, leading to damage by accumulation of a toxic substrate or deficiency of an essential metabolite. Early diagnosis is crucial in many of these conditions to prevent or minimise brain damage. Whilst many of the neuroimaging features are nonspecific, certain disorders demonstrate specific patterns due to selective vulnerability of different structures to different insults.