Testing Darwin's Hypothesis on the Evolution of Ornamental Eyespots In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Molecular Phylogeny of the Peacock-Pheasants (Galliformes: Polyplectron Spp.) Indicates Loss and Reduction of Ornamental Traits and Display Behaviours
Biological Journal of the Linnean Society (2001), 73: 187–198. With 3 figures doi:10.1006/bijl.2001.0536, available online at http://www.idealibrary.com on A molecular phylogeny of the peacock-pheasants (Galliformes: Polyplectron spp.) indicates loss and reduction of ornamental traits and display behaviours REBECCA T. KIMBALL1,2∗, EDWARD L. BRAUN1,3, J. DAVID LIGON1, VITTORIO LUCCHINI4 and ETTORE RANDI4 1Department of Biology, University of New Mexico, Albuquerque, NM 87131, USA 2Department of Evolution, Ecology and Organismal Biology, Ohio State University, Columbus, OH 43210, USA 3Department of Plant Biology, Ohio State University, Columbus, OH 43210, USA 4Istituto Nazionale per la Fauna Selvatica, via Ca` Fornacetta 9, 40064 Ozzano dell’Emilia (BO), Italy Received 4 September 2000; accepted for publication 3 March 2001 The South-east Asian pheasant genus Polyplectron is comprised of six or seven species which are characterized by ocelli (ornamental eye-spots) in all but one species, though the sizes and distribution of ocelli vary among species. All Polyplectron species have lateral displays, but species with ocelli also display frontally to females, with feathers held erect and spread to clearly display the ocelli. The two least ornamented Polyplectron species, one of which completely lacks ocelli, have been considered the primitive members of the genus, implying that ocelli are derived. We examined this hypothesis phylogenetically using complete mitochondrial cytochrome b and control region sequences, as well as sequences from intron G in the nuclear ovomucoid gene, and found that the two least ornamented species are in fact the most recently evolved. Thus, the absence and reduction of ocelli and other ornamental traits in Polyplectronare recent losses. -

Document Resume Ed 049 958 So 000 779 Institution Pub
DOCUMENT RESUME ED 049 958 SO 000 779 AUTHCE Nakosteen, Mehdi TITLE Conflicting Educational Ideals in America, 1775-1831: Documentary Source Book. INSTITUTION Colorado Univ., Boulder. School of Education. PUB DATE 71 NOTE 480p. EDES PRICE EDES Price MF-SC.65 HC-$16.45 DESCRIPTORS *Annotated Bibliographies, Cultural Factors, *Educational History, Educational Legislation, *Educational Practice, Educational Problems, *Educational Theories, Historical Reviews, Resource Materials, Social Factors, *United States History IDENTIFIERS * Documentary History ABSTRACT Educational thought among political, religious, educational, and other social leaders during the formative decades of American national life was the focus of the author's research. The initial objective was the discovery cf primary materials from the period to fill a gap in the history of American educational thought and practice. Extensive searching cf unpublished and uncatalogued library holdings, mainly those of major public and university libraries, yielded a significant quantity of primary documents for this bibliography. The historical and contemporary works, comprising approximately 4,500 primary and secondary educational resources with some surveying the cultural setting of educational thinking in this period, are organized around 26 topics and 109 subtopics with cross-references. Among the educational issues covered by the cited materials are: public vs. private; coed vs. separate; academic freedom, teacher education; teaching and learning theory; and, equality of educational opportunity. In addition to historical surveys and other secondary materials, primary documents include: government documents, books, journals, newspapers, and speeches. (Author/DJB) CO Lir\ 0 CY% -1- OCY% w CONFLICTING EDUCATIONAL I D E A L S I N A M E R I C A , 1 7 7 5 - 1 8 3 1 : DOCUMENTARY SOURCE B 0 0 K by MEHDI NAKOSTEEN Professor of History and Philosophy of Education University of Colorado U.S. -

Hybridization & Zoogeographic Patterns in Pheasants
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Paul Johnsgard Collection Papers in the Biological Sciences 1983 Hybridization & Zoogeographic Patterns in Pheasants Paul A. Johnsgard University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/johnsgard Part of the Ornithology Commons Johnsgard, Paul A., "Hybridization & Zoogeographic Patterns in Pheasants" (1983). Paul Johnsgard Collection. 17. https://digitalcommons.unl.edu/johnsgard/17 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Paul Johnsgard Collection by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. HYBRIDIZATION & ZOOGEOGRAPHIC PATTERNS IN PHEASANTS PAUL A. JOHNSGARD The purpose of this paper is to infonn members of the W.P.A. of an unusual scientific use of the extent and significance of hybridization among pheasants (tribe Phasianini in the proposed classification of Johnsgard~ 1973). This has occasionally occurred naturally, as for example between such locally sympatric species pairs as the kalij (Lophura leucol11elana) and the silver pheasant (L. nycthelnera), but usually occurs "'accidentally" in captive birds, especially in the absence of conspecific mates. Rarely has it been specifically planned for scientific purposes, such as for obtaining genetic, morphological, or biochemical information on hybrid haemoglobins (Brush. 1967), trans ferins (Crozier, 1967), or immunoelectrophoretic comparisons of blood sera (Sato, Ishi and HiraI, 1967). The literature has been summarized by Gray (1958), Delacour (1977), and Rutgers and Norris (1970). Some of these alleged hybrids, especially those not involving other Galliformes, were inadequately doculnented, and in a few cases such as a supposed hybrid between domestic fowl (Gallus gal/us) and the lyrebird (Menura novaehollandiae) can be discounted. -

Argus 2019 Q1
The Argus January 2019 A.S. LIII _____________________________________________________ Barony of One Thousand Eyes, Kingdom of Artemisia “The Heart of Artemisia” This is theJanuary 2019 issue of the Argus, a publication of the Barony of one Thousand Eyes, Kingdom of Artemesia, Society for Creative Anachronism, Inc. and is available from Lisa Edmiston, [email protected]. It is not a corporate publication of the Society for Creative Anachronism, Inc. and does not delineate SCA, Inc. policies. Copyright 2019, Society for Creative Anachronism Inc. For information on reprinting photographs, articles, or artwork from this publication please contact the Chronicler, who will assist you in contacting the original creator of the piece. Please respect the legal rights of our contributors. Join The Current Middle Ages! The Society for Creative Anachronism, or SCA, is an international organization dedicated to researching and re-creating the arts, skills, and traditions of pre-17th-century Europe. Members of the SCA study and take part in a variety of activities, including combat, archery, equestrian activities, costuming, cooking, metalwork, woodworking, music, dance, calligraphy, fiber arts, and much more. If it was done in the Middle Ages or Renaissance, odds are you’ll find someone in the SCA interested in recreating it. You will frequently hear SCA participants describe the SCA as recreating the Middle Ages “as they ought to have been.” In some ways this is true – we choose to use indoor plumbing, heated halls, and sewing -

Phylogenetic Relationships of the Phasianidae Reveals Possible Non-Pheasant Taxa
Journal of Heredity 2003:94(6):472–489 Ó 2003 The American Genetic Association DOI: 10.1093/jhered/esg092 Phylogenetic Relationships of the Phasianidae Reveals Possible Non-Pheasant Taxa K. L. BUSH AND C. STROBECK From the Department of Biological Sciences, University of Alberta–Edmonton, Edmonton, Alberta T6G 2E9, Canada. Address correspondence to Krista Bush at the address above, or e-mail: [email protected]. Abstract The phylogenetic relationships of 21 pheasant and 6 non-pheasant species were determined using nucleotide sequences from the mitochondrial cytochrome b gene. Maximum parsimony and maximum likelihood analysis were used to try to resolve the phylogenetic relationships within Phasianidae. Both the degree of resolution and strength of support are improved over previous studies due to the testing of a number of species from multiple pheasant genera, but several major ambiguities persist. Polyplectron bicalcaratum (grey peacock pheasant) is shown not to be a pheasant. Alternatively, it appears ancestral to either the partridges or peafowl. Pucrasia macrolopha macrolopha (koklass) and Gallus gallus (red jungle fowl) both emerge as non-pheasant genera. Monophyly of the pheasant group is challenged if Pucrasia macrolopha macrolopha and Gallus gallus are considered to be pheasants. The placement of Catreus wallichii (cheer) within the pheasants also remains undetermined, as does the cause for the great sequence divergence in Chrysolophus pictus obscurus (black-throated golden). These results suggest that alterations in taxonomic -
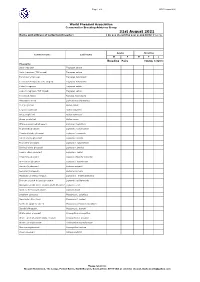
31St August 2021 Name and Address of Collection/Breeder: Do You Closed Ring Your Young Birds? Yes / No
Page 1 of 3 WPA Census 2021 World Pheasant Association Conservation Breeding Advisory Group 31st August 2021 Name and address of collection/breeder: Do you closed ring your young birds? Yes / No Adults Juveniles Common name Latin name M F M F ? Breeding Pairs YOUNG 12 MTH+ Pheasants Satyr tragopan Tragopan satyra Satyr tragopan (TRS ringed) Tragopan satyra Temminck's tragopan Tragopan temminckii Temminck's tragopan (TRT ringed) Tragopan temminckii Cabot's tragopan Tragopan caboti Cabot's tragopan (TRT ringed) Tragopan caboti Koklass pheasant Pucrasia macrolopha Himalayan monal Lophophorus impeyanus Red junglefowl Gallus gallus Ceylon junglefowl Gallus lafayettei Grey junglefowl Gallus sonneratii Green junglefowl Gallus varius White-crested kalij pheasant Lophura l. hamiltoni Nepal Kalij pheasant Lophura l. leucomelana Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata True silver pheasant Lophura n. nycthemera Berlioz’s silver pheasant Lophura n. berliozi Lewis’s silver pheasant Lophura n. lewisi Edwards's pheasant Lophura edwardsi edwardsi Vietnamese pheasant Lophura e. hatinhensis Swinhoe's pheasant Lophura swinhoii Salvadori's pheasant Lophura inornata Malaysian crestless fireback Lophura e. erythrophthalma Bornean crested fireback pheasant Lophura i. ignita/nobilis Malaysian crestless fireback/Vieillot's Pheasant Lophura i. rufa Siamese fireback pheasant Lophura diardi Southern Cavcasus Phasianus C. colchicus Manchurian Ring Neck Phasianus C. pallasi Northern Japanese Green Phasianus versicolor -

The Argus Pheasant Is at Times One of the Most Ornamental Member of the En Tire Pheasant Family
c ~ CJ) :> > () c ro Z > .D C/) o .!:: 0.. by Francis Billie The Argus Pheasant is at times one of the most ornamental member of the en tire Pheasant Family. Adult males can measure ix to even feet from the tip of the beak to the tip of the tail, although most of this length is taken up by their tail feather . In coloration the Argus is rather dull and plain, it is only during its marvelous courtship display that it manifest its great beauty and elaborate markings. There are two genera of Argus each containing two subspecies. In the genus Rehinartia there is Rheinart's Crested Close-up feather detads of the Arj?us Pheasant. Argus Pheasant, male photos by Francis Billie Argus Pheasant, hen 38 Argus, (Rheinartia ocellata ocellata), back and forth and he vibrates his found in Vietnam and parts of Laos, and plumage so the ocelli appear to revolve. the Malay Crested Argus (Rheinartia During this display the male's head is hid ocellalla nigrescens) a rare, little known den from view. bird found in various parts of the Malay It is interesting to note that the Argus Peninsula. At present, the status of the display is not like that of the Peacock bird in Vietnam is unknown, but the Pheasants in that the frontal display is not made with the tail feathers but is formed ,c F P r I Malayan subspecies has been observed in '{;HI \ t ') the wild state during a recent expedition in with the wings. It is also interesting that the f'PICrS & 81RUS the area. -

Illegal Wildlife Trade 02
2019 02 Illegal Wildlife Trade Wildlife Illegal NEWS 1 WAZA Executive Office Staff Interim CEO and Martín Zordan Director of Conservation [email protected] Chief Operating Officer Christina Morbin [email protected] Director of Communications Gavrielle Kirk-Cohen [email protected] Director of Membership Janet Ho [email protected] Animal Welfare and Paula Cerdán Conservation Coordinator [email protected] Imprint WAZA Executive Office Contacts Postal Address WAZA Executive Office Editor: Carrer de Roger de Llúria 2, 2-2 Gavrielle Kirk-Cohen 08010 Barcelona Spain Proofreader Phone +34 936638811 Laurie Clinton Email [email protected] Website www.waza.org Layout and design: Facebook www.facebook.com/officialWAZA Butterhalfsix.com Twitter twitter.com/WAZA This edition of WAZA News is Instagram www.instagram.com/wazaglobal also available at: www.waza.org WAZA Membership Printed on FSC-certified paper WAZA members as of 24 October 2019 Affiliates 10 Associations 24 Corporates 19 Institutions 282 Future Events 2020: WAZA/IZE Annual Conference, San Diego Zoo, San Diego, USA 11-15 October Cover Photo: Young chimpanzee : Moscow Zoo, Moscow, Russia rescued from the illegal wildlife trade. 2021 ©️ Pan African Sanctuary Alliance (PASA) 2022: Loro Parque, Tenerife, Spain President’s Letter Jenny Gray WAZA President July 2017 - November 2019 It is hard to isolate a single threat as contributing to the local extinction of species in the wild. Most often, various different threats overlap, each one compounding the negative impacts and ultimately resulting in the collapse of a species. Yet, I can’t help thinking that the illegal removal and trade of animals must be the most evil of the contributing factors. -

Remembering Dr. Arthur Crane Risser by Josef Lindholm III, It Stayed That Way Through the Rest of the 1960’S
Remembering Dr. Arthur Crane Risser by Josef Lindholm III, it stayed that way through the rest of the 1960’s. On Dec. 31, Senior Aviculturist, The Dallas World Aquarium 1969, it reached an all-time high of 1,126 species and subspecies of birds (and 3,465 specimens). Then it dropped. At the end of Art Risser’s death following a stroke on the day after Christ- 1970 there were 1,097 taxa. On Jan. 1, 1972, there were 917. A mas 2008, was entirely unexpected. But many of his saddened year later there were 856. And on Jan1., 1974, the number stood friends were also startled to learn he was 70. I think most of us at 772. I found this deeply disturbing. thought he was far younger. When I first met him, shortly after At the same time, my own small avicultural world had also his arrival at the San Diego Zoo, as Assistant Curator of Birds, become much smaller. In 1972, I was, with much effort, able to in 1974, I thought he was in his late twenties. He was, in fact, 35 convince my parents to buy me Red-eared Waxbills at Wool- when he thus entered the zoo profession, having previously been worth’s and Strawberry Finches and Cut-throats at the White involved in mammalogy. Front, all for $3.95 a pair. In 1974, I found the prices for all of He earned his Master’s in Wildlife Management from the these were now $40 a pair. University of Arizona, in 1963, conducting field research on In answer to the question that all young zoo enthusiasts ask: White-nosed Coatis. -

Gamebird Conservation
Threatened Ducks and Geese West Indian Whistling Duck Hawaiian Goose Gamebird Conservation Freckled Duck by Jack Clinton-Eitniear Crested Shelduck San Antonio, Texas Baykal Teal New Zealand Brown Teal Laysan Duck Pink-headed Duck Madagascar Pochard During the first half of the nine from the list at the end of this article, Scaly-sided Merganser teenth century, had you ventured into they have a rather formidable chal Lesser White-fronted Goose a meat market in ew York or Balti lenge ahead of them. Red-breasted Goose more, you might well have encoun While it is encouraging to note that Ruddy-headed Goose tered a rather "fishy" tasting duck a number of species facing troubles in White-winged Duck offered for sale. That duck, often said the wild are well represented in cap Madagascar Teal to have rotted as few desired to eat tivity it is equally saddening that Hawaiian Duck them, was that of a Labrador Duck most are not. Having had the pleasure Marbled Teal (Camptorhynchus labradorius)~ a of working with tragopans, as well as Baer's Pochard now extinct species. While dis brown-, blue- and white-eared pheas Brazilian Merganser covered inhabiting the northeast sea ants in the late sixties, I know well of White-headed Duck board in 1789, the duck, for reasons their awe-inspiring beauty. Anyone unknown, had disappeared by 1878. who is tempted to associate bright Threatened Pheasants, While at least four species of water colors of enchanting combinations Francolins, Quail & Peafowl fowl have passed into extinction, I with only small birds needs to visit a Bearded Wood-partridge know of only one Gallinaceous bird, pheasant collection. -

<I>ARGUSIANUS ARGUS</I>
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Natural Resources Natural Resources, School of 2009 MOVEMENTS, DISTRIBUTION, AND ABUNDANCE OF GREAT ARGUS PHEASANTS (ARGUSIANUS ARGUS) IN A SUMATRAN RAINFOREST Nurul L. Winarni University of Georgia Timothy G. O'Brien Wildlife Conservation Society, [email protected] John P. Carroll University of Georgia, [email protected] Margaret F. Kinnaird Wildlife Conservation Society Follow this and additional works at: http://digitalcommons.unl.edu/natrespapers Part of the Natural Resources and Conservation Commons, Natural Resources Management and Policy Commons, and the Other Environmental Sciences Commons Winarni, Nurul L.; O'Brien, Timothy G.; Carroll, John P.; and Kinnaird, Margaret F., "MOVEMENTS, DISTRIBUTION, AND ABUNDANCE OF GREAT ARGUS PHEASANTS (ARGUSIANUS ARGUS) IN A SUMATRAN RAINFOREST" (2009). Papers in Natural Resources. 658. http://digitalcommons.unl.edu/natrespapers/658 This Article is brought to you for free and open access by the Natural Resources, School of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Natural Resources by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. The Auk 126(2):341 350, 2009 The American Ornithologists’ Union, 2009. Printed in USA. MOVEMENTS, DISTRIBUTION, AND ABUNDANCE OF GREAT ARGUS PHEASANTS (ARGUSIANUS ARGUS) IN A SUMATRAN RAINFOREST NURUL L. WINARNI,1,2 TIMOTHY G. O’BRIEN,3,4,5 JOHN P. C ARROLL,2 AND MARGARET F. K INNAIRD3,4 1Wildlife Conservation Society, Indonesia Program, Jl. Ciremei 8, P.O. Box 311, Bogor 16003, Indonesia; 2D.B. Warnell School of Forest Resources, University of Georgia, Athens, Georgia 30605, USA; 3Wildlife Conservation Society, 2300 Southern Boulevard, Bronx, New York 10460, USA; and 4Mpala Research Center, P.O. -

Academy of Natural Sciences of Drexel Uni- Altissima, Ailanthus 48, 49, 57, 62, 181, 182 Versity 45, 77, 89, 111, 151, 178, 198, Amaranthus 26 247, 276
alkaloids 293 Allee effects 178, 179 Index Allee, Warder Clyde 172, 178 Allegheny River 301 A Allentown, Pennsylvania 271, 272, 274 altissima, Ageratina 55 Academy of Natural Sciences of Drexel Uni- altissima, Ailanthus 48, 49, 57, 62, 181, 182 versity 45, 77, 89, 111, 151, 178, 198, Amaranthus 26 247, 276. See also Academy of Natural Amaranthus hybridus 184 Sciences of Philadelphia Amaranthus retroflexus 180 Academy of Natural Sciences of Philadelphia 2, Ambigolimax valentianus 172 35, 48, 53, 68, 78, 87, 111, 118, 155, 164, Ambrosia 26 168, 179, 206, 208, 212, 216, 230, 300 Ambrosia artemisiifolia 203, 296 Acanalonia conica 228 Ameiurus catus 216 Acanthocephala 168–170 Ameiurus nebulosus 215–226. See also brown “An Account of the Crustacea of the United bullhead catfish States” 164 americana, Malacosoma 51 Acer 78 americana, Phytolacca 140–142 acerifolia, Platanus 5 americana, Setophaga [Parula] 67–74. See Acer negundo 314 also parula warblers Acer palmatum 41 American Civic Association 243 Acer platanoides 247 American crow 150 acid rain 299 American Ornithologist’s Union 70 Acrelius, Israel 128, 130 American Ornithology 9, 140 adventitium, Bipalium 136 American Philosophical Society of Philadelphia Aedes aegypti 307 48, 174, 238 Aedes albopictus 307 American robin 138, 139–149, 298, 301, 303, aegypti, Aedes 307 306 Aesculus sp. 16 American Spiders and Their Spinning Work 96 Ageratina altissima 55 American toad 230 aggregation behavior americanus, Bufo 230 agricultural pests 274, 275. See also specific American Water Works Association 301 pests Ampelopsis brevipedunculata 117, 123 Ailanthus altissima 48, 49, 57, 62, 181, 182 Amphibia 230 ailanthus silkmoth 45–52, 316 amphibians, urban dryness and 230–231 ailanthus tree 20, 28, 46–47, 50, 53, 57, 62, 63, Amphidasys betularia 245.