Computer Vision News the Magazine of the Algorithm Community
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Desktop (Overview)
The desktop (overview) The desktop is the main screen area that you see after you turn on your computer and log on to Windows. Like the top of an actual desk, it serves as a surface for your work. When you open programs or folders, they appear on the desktop. You can also put things on the desktop, such as files and folders, and arrange them however you want. The desktop is sometimes defined more broadly to include the taskbar and Windows Sidebar. The taskbar sits at the bottom of your screen. It shows you which programs are running and allows you to switch between them. It also contains the Start button , which you can use to access programs, folders, and computer settings. On the side of the screen, Sidebar contains small programs called gadgets. The desktop, taskbar, and Sidebar Where did my desktop go? Because programs run on top of the desktop, the desktop itself is often partially or completely hidden. But it's still there, underneath everything. To see the whole desktop without closing any of your open programs or windows, click the Show Desktop button on the taskbar. The desktop is revealed. Click the icon again to restore all of your windows to the way they were. Desktop Training Session Handout Page 1 http://ict.maxwell.syr.edu/vista/ Working with desktop icons Icons are small pictures that represent files, folders, programs, and other items. When you first start Windows, you'll see at least one icon on your desktop: the Recycle Bin (more on that later). -

Bforartists UI Redesign Design Document Part 2 - Theming
Bforartists UI redesign Design document part 2 - Theming Content Preface...........................................................................................................................6 The editor and window types......................................................................................7 Python console.............................................................................................................8 Layout:................................................................................................................................................................8 The Console Window.........................................................................................................................................8 Menu bar with a menu........................................................................................................................................8 Dropdown box with icon....................................................................................................................................9 RMB menu for menu bar....................................................................................................................................9 Toolbar................................................................................................................................................................9 Button Textform..................................................................................................................................................9 -

Navigate Windows and Folders
Windows® 7 Step by Step by Joan Preppernau and Joyce Cox To learn more about this book, visit Microsoft Learning at http://www.microsoft.com/MSPress/books/ 9780735626676 ©2009 Joan Preppernau and Joyce Cox Early Content—Subject to Change Windows 7 Step by Step Advance Content–Subject to Change Windows 7 Step by Step Draft Table of Contents Overview Front Matter ............................................................................................................................................. 3 Contents ................................................................................................................................................ 3 About the Authors ................................................................................................................................ 3 Features and Conventions of This Book ................................................................................................ 3 Using the Companion CD ...................................................................................................................... 3 Getting Help .......................................................................................................................................... 3 Introducing Windows 7 ......................................................................................................................... 3 Part I: Getting Started with Windows 7 .................................................................................................... 4 1 Explore Windows 7 ........................................................................................................................... -

Editing the Home Screen Widgets:
Brisbane German Club WordPress Website Manual 1 Table of Contents How To Add: A Page .............................................................................................................. 3 How To Edit: A Page .............................................................................................................. 4 How To Add: A Post ............................................................................................................... 8 How To Edit: A Post ............................................................................................................. 12 How To Edit: Home Page ..................................................................................................... 13 Editing the Home Screen Widgets: ....................................................................................... 19 How To Change: The Menu .................................................................................................. 22 Events & Calendar ............................................................................................................... 24 How To Add: a Hyperlink ..................................................................................................... 26 2 How To Add: A Page Step 1: Go to „Pages‟ on the left sidebar of the backend. Click „Add New‟ 3 How To Edit: A Page Step 1: Hold the cursor over any specific page. 4 options appear – to edit/ quick edit/ trash/ view. These are self-explanatory however to edit a page select the edit function or alternatively just click on -
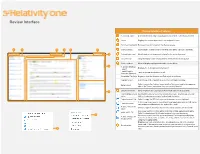
Review Interface Quick Reference
Review Interface Review Interface Features Viewer type tabs Select which Viewer type you would like to view the current document in. Viewer Displays the current document in the selected Viewer. Document navigation Browse across documents in the Review queue. 9 6 8 1 2 3 Coding layouts Customizable coding forms for viewing and editing a document’s fields. 7 Related items card Identify and act on documents related to the active document. Document list List presentation of all the documents in the user’s Review queue. Viewer settings Menu of display configurations and review settings. 4 Keyboard Shortcut Displays the keyboard shortcuts legend. Legend Enable/Disable Turns keyboard shortcuts on or off. Keyboard Shortcuts Show/Hide Tab Strip Display or hide the Sidebar and Tab strip in the Viewer. Pop Out Viewer Select to open the Pop Out Viewer in a new browser window. Select to move the Coding Layout and the Document card to the opposite Swap Layout side of the Viewer from where they are currently located. Document Actions Menu of actions the user may perform on the document as a whole. 5 Download document Download the native file for the current document. Alternatively, you can native click on the document’s name to download the native. Copy document link Select to copy the URL to the current document to your clipboard. Select to open an email in your default email application with the URL to the Email document current document included in the body of the email. Replace document Select to replace the native file of the current document with a new file native Hover your cursor over this option and Image Profile options will display. -

Vigil Client 10 | User's Guide
Client 10 Remote Desktop Client Software User Guide Revised: June 1st, 2017 3xLOGIC Inc. ©2017 COPYRIGHT AND LEGAL NOTICES|VIGIL CLIENT 10 | USER'S GUIDE VIGIL Copyright ©2017 3xLOGIC, Inc. All rights reserved. 3xLOGIC Inc. 210-10385 Westmoor Drive Westminster, CO 80021 United States (303) 430-1969 Disclaimer Information in this document is subject to change without notice and does not represent a commitment on the part of 3xLOGIC Inc. The soft- ware and /or databases described in this document are furnished under a license agreement or nondisclosure agreement. They may be used or copied only in accordance with the terms of the agreement. It is against the law to copy the software on any medium except as specifically allowed in the license or nondisclosure agreement. The purchaser may make one copy of the software for backup purposes. No part of this manual and /or databases may be reproduced or transmitted in any form or by any means, electronic or mechanical, including (but not limited to) photocopying, recording, or information storage and retrieval systems, for any purpose other than the purchaser's personal use, without the express written permission of 3xLOGIC Inc. 3xLOGIC, VIGIL and AZTECH are trademarks of 3xLOGIC, Inc. Other trademarks and trade names may be used in this document to refer to either the entities claiming the marks and names or their products. 3xLOGIC Inc. disclaim any proprietary interest in trademarks and trade names other than their own. TABLE OF CONTENTS|VIGIL CLIENT 10 | USER'S GUIDE Table of Contents 1 Introduction -

Microsoft Office 2016! Step
spine = 0.8291” The quick way to get started with Microsoft Office 2016! Step Covers Microsoft Word, Excel, PowerPoint, and Outlook by Office 2016 Microsoft Step Microsoft This is learning made easy. Get more done quickly with IN FULL COLOR! Microsoft Word, Excel, PowerPoint, and Outlook. Jump in wherever you need answers—brisk lessons and Office 2016 colorful screenshots show you exactly what to do, step by step. • Format documents for visual impact • Quickly prepare personalized email messages and labels • Build powerful workbooks for analysis and reporting • Analyze alternative data sets with Quick Analysis Lens, Goal Seek, and Solver • Prepare highly effective presentations • Strengthen your presentations by adding tables and graphics • Organize your email, scheduling, and contacts • Look up just the tasks and lessons you need Step Colorful screenshots by Step Download your Step by Step practice files at: Helpful tips and http://aka.ms/Office2016sbs/downloads pointers Easy numbered Lambert Frye steps MicrosoftPressStore.com ISBN 978-0-7356-9923-6 U.S.A. $44.99 44999 Canada $55.99 [Recommended] 9 780735 699236 Microsoft Office Joan Lambert and Curtis Frye PRACTICE FILES Celebrating over 30 years! 9780735699236_Office2016SBS_cover.indd 1 10/29/2015 12:13:47 PM Microsoft Office 2016 Step by Step Joan Lambert Curtis Frye 699236_Office2016SBS.indb 1 10/29/2015 6:33:02 PM PUBLISHED BY Microsoft Press A division of Microsoft Corporation One Microsoft Way Redmond, Washington 98052-6399 Copyright © 2015 by Curtis Frye and Joan Lambert All rights reserved. No part of the contents of this book may be reproduced or transmitted in any form or by any means without the written permission of the publisher. -

Toolbar,Sidebar and Statusbar Libreoffice (Writer)
NIELIT Gorakhpur Course Name: O Level (1st Sem) Subject: ITTNB Topic: Toolbar,Sidebar and Statusbar Date: 21-04-20 Libreoffice [Writer] Toolbar:- The toolbar, also called bar or standard toolbar, is a row of boxes, often at the top of an application window, that control various functions of the software. The boxes often contain images that correspond with the function they control. A toolbar often provides quick access to functions that are commonly performed in the program. For example, a formatting toolbar gives us access to things like making text bold or changing its alignment, along with other common buttons. In an Internet browser, toolbars add functionality that may not come pre-installed. For example, with the Google toolbar, you can get access to exclusive Google features. To activate it, enable View > Toolbars > Standard (Single Mode) and disable View > Toolbars > Standard and View > Toolbars > Formatting. Sidebar:-The Sidebar is located on the right side of the edit views of Writer, Calc, Impress, and Draw. It contains one or more panels, based on the current document context. Panels are organized into decks. A tab bar on the right side of the sidebar allows you to switch between different decks. All components contain the Properties, Styles and Formatting, Pages , Gallery, and Navigator decks. Some components have additional decks, such as Master Pages, Custom Animation, and Slide Transition for Impress; Manage Changes for Writer; and Functions for Calc. To activate the Sidebar, select View > Sidebar from the Menu bar. To hide the Sidebar, click on the gray Hide button on the left. Click on the same button to show the Sidebar again. -

Explore Windows 7
Windows® 7 Step by Step by Joan Preppernau and Joyce Cox To learn more about this book, visit Microsoft Learning at http://www.microsoft.com/MSPress/books/ 9780735626676 ©2009 Joan Preppernau and Joyce Cox Early Content—Subject to Change Windows 7 Step by Step Advance Content–Subject to Change Windows 7 Step by Step Draft Table of Contents Overview Front Matter ............................................................................................................................................. 3 Contents ................................................................................................................................................ 3 About the Authors ................................................................................................................................ 3 Features and Conventions of This Book ................................................................................................ 3 Using the Companion CD ...................................................................................................................... 3 Getting Help .......................................................................................................................................... 3 Introducing Windows 7 ......................................................................................................................... 3 Part I: Getting Started with Windows 7 .................................................................................................... 4 1 Explore Windows 7 ........................................................................................................................... -

Microsoft Word 2011
Microsoft Word 2011 Prepared by Computing Services at the Eastman School of Music – May 2011 Table of Contents New Look in Microsoft Office 2011 ................................................................................................................................... 4 Standard Toolbar ........................................................................................................................................................... 4 Ribbon ............................................................................................................................................................................ 4 Appearance of Microsoft Word ......................................................................................................................................... 5 Creating a New Document ................................................................................................................................................. 6 Opening a Document ........................................................................................................................................................ 6 Saving a Document ............................................................................................................................................................ 7 Home Tab – Shaping your Document ................................................................................................................................ 8 Copy/Paste Text ............................................................................................................................................................ -
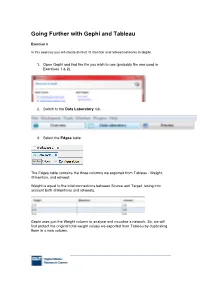
Going Further with Gephi and Tableau
Going Further with Gephi and Tableau Exercise 3 In this exercise you will create distinct @ mention and retweet networks in Gephi. 1. Open Gephi and find the file you wish to use (probably the one used in Exercises 1 & 2). 2. Switch to the Data Laboratory tab. 3. Select the Edges table. The Edges table contains the three columns we exported from Tableau - Weight, @mention, and retweet: Weight is equal to the total connections between Source and Target, taking into account both @mentions and retweets. Gephi uses just the Weight column to analyse and visualise a network. So, we will first protect the original total weight values we exported from Tableau by duplicating them in a new column. 4. In the button bar at the bottom of the Gephi window, select Duplicate Column. 5. Click on Weight to select the column you want to duplicate. 6. Name the new column Weight (original), and click OK. The Edges table now contains a new column that duplicates Weight. 7. We can now safely reset the Weight values to zero. In the button bar at the bottom, click on Fill column with a value. 8. Select Weight. 9. Enter 0 and click OK . You’ve now reset all edge weights to zero. 10. We’ll now focus first on the retweet network. In the button bar at the bottom, click on Copy data to other columns. 11. Select retweet. 12. Choose Weight as the destination column and click OK. The Weight column now matches the value of retweet, with zeros where there were no retweets between two accounts. -

Cascade Training Transcript
Cascade Training Table of Contents Step 1: Download the Files ......................................................................................................................................................... 3 Step 2: Login to Cascade ............................................................................................................................................................ 3 Step 3: Learn how to Navigate Cascade ................................................................................................................................ 3 The Dashboard: ........................................................................................................................................................................ 3 The Menu Bar ............................................................................................................................................................................ 4 Site List Explanation ................................................................................................................................................................. 4 The Asset Tree ........................................................................................................................................................................... 4 The Sub Menu ............................................................................................................................................................................ 5 Step 4: The Templates Explained .............................................................................................................................................