Recurrent Somatic MAP2K1 Mutations in Papillary Thyroid Cancer and Colorectal Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Map2k1 and Map2k2 Genes Contribute to the Normal Development of Syncytiotrophoblasts During Placentation
RESEARCH ARTICLE 1363 Development 136, 1363-1374 (2009) doi:10.1242/dev.031872 Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation Valérie Nadeau*, Stéphanie Guillemette*, Louis-François Bélanger, Olivier Jacob, Sophie Roy and Jean Charron† The mammalian genome contains two ERK/MAP kinase kinase genes, Map2k1 and Map2k2, which encode dual-specificity kinases responsible for ERK/MAP kinase activation. In the mouse, loss of Map2k1 function causes embryonic lethality, whereas Map2k2 mutants survive with a normal lifespan, suggesting that Map2k1 masks the phenotype due to the Map2k2 mutation. To uncover the specific function of MAP2K2 and the threshold requirement of MAP2K proteins during embryo formation, we have successively ablated the Map2k gene functions. We report here that Map2k2 haploinsufficiency affects the normal development of placenta in the absence of one Map2k1 allele. Most Map2k1+/–Map2k2+/– embryos die during gestation because of placenta defects restricted to extra-embryonic tissues. The impaired viability of Map2k1+/–Map2k2+/– embryos can be rescued when the Map2k1 deletion is restricted to the embryonic tissues. The severity of the placenta phenotype is dependent on the number of Map2k mutant alleles, the deletion of the Map2k1 allele being more deleterious. Moreover, the deletion of one or both Map2k2 alleles in the context of one null Map2k1 allele leads to the formation of multinucleated trophoblast giant (MTG) cells. Genetic experiments indicate that these structures are derived from Gcm1-expressing syncytiotrophoblasts (SynT), which are affected in their ability to form the uniform SynT layer II lining the maternal sinuses. Thus, even though Map2k1 plays a predominant role, these results enlighten the function of Map2k2 in placenta development. -

The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma
CD-13-0617_PAP.indd Page OF1 19/11/13 9:36 PM user-f028 /Books-Arts/JOURNAL-Cancer%20Discovery/01-JAN-Issue-2014/PAP Published OnlineFirst November 21, 2013; DOI: 10.1158/2159-8290.CD-13-0617 RESEARCH ARTICLE The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma Eliezer M. Van Allen 1 , 3 , Nikhil Wagle 1 , 3 , Antje Sucker 5 , 6 , Daniel J. Treacy 1 , Cory M. Johannessen 3 , Eva M. Goetz 1 , Chelsea S. Place 1 , 3 , Amaro Taylor-Weiner 3 , Steven Whittaker 3 , Gregory V. Kryukov 3 , Eran Hodis 1 , 3,4 , Mara Rosenberg 3 , Aaron McKenna 3 , 15 , Kristian Cibulskis 3 , Deborah Farlow 3 , Lisa Zimmer 5 , 6 , Uwe Hillen 5 , 6 , Ralf Gutzmer 8 , Simone M. Goldinger 16 , Selma Ugurel 9 , Helen J. Gogas 17 , Friederike Egberts 10 , Carola Berking 6 , 11 , Uwe Trefzer 6 , 12 , Carmen Loquai 6 , 13 , Benjamin Weide 6 , 14 , Jessica C. Hassel 6 , 7 , Stacey B. Gabriel 3 , Scott L. Carter 3 , Gad Getz 2 , 3 , Levi A. Garraway 1 , 3 , and Dirk Schadendorf 5 , 6 on behalf of the Dermatologic Cooperative Oncology Group of Germany (DeCOG) Downloaded from cancerdiscovery.aacrjournals.org on September 25, 2021. © 2013 American Association for Cancer Research. CD-13-0617_PAP.indd Page OF2 19/11/13 9:36 PM user-f028 /Books-Arts/JOURNAL-Cancer%20Discovery/01-JAN-Issue-2014/PAP Published OnlineFirst November 21, 2013; DOI: 10.1158/2159-8290.CD-13-0617 ABSTRACT Most patients with BRAF V600 -mutant metastatic melanoma develop resistance to selective RAF kinase inhibitors. The spectrum of clinical genetic resistance mechanisms to RAF inhibitors and options for salvage therapy are incompletely understood. -

Personalized Prediction of Acquired Resistance to EGFR-Targeted Inhibitors Using a Pathway-Based Machine Learning Approach
Cancers 2019, 11, x S1 of S9 Supplementary Materials: Personalized Prediction of Acquired Resistance to EGFR-Targeted Inhibitors Using a Pathway-Based Machine Learning Approach Young Rae Kim, Yong Wan Kim, Suh Eun Lee, Hye Won Yang and Sung Young Kim Table S1. Characteristics of individual studies. Sample Size Origin of Cancer Drug Dataset Platform S AR (Cell Lines) Lung cancer Agilent-014850 Whole Human GSE34228 26 26 (PC9) Genome Microarray 4x44K Gefitinib Epidermoid carcinoma Affymetrix Human Genome U133 GSE10696 3 3 (A431) Plus 2.0 Head and neck cancer Illumina HumanHT-12 V4.0 GSE62061 12 12 (Cal-27, SSC-25, FaDu, expression beadchip SQ20B) Erlotinib Head and neck cancer Illumina HumanHT-12 V4.0 GSE49135 3 3 (HN5) expression beadchip Lung cancer (HCC827, Illumina HumanHT-12 V3.0 GSE38310 3 6 ER3, T15-2) expression beadchip Lung cancer Illumina HumanHT-12 V3.0 GSE62504 1 2 (HCC827) expression beadchip Afatinib Lung cancer * Illumina HumanHT-12 V4.0 GSE75468 1 3 (HCC827) expression beadchip Head and neck cancer Affymetrix Human Genome U133 Cetuximab GSE21483 3 3 (SCC1) Plus 2.0 Array GEO, gene expression omnibus; GSE, gene expression series; S, sensitive; AR, acquired EGFR-TKI resistant; * Lung Cancer Cells Derived from Tumor Xenograft Model. Table S2. The performances of four penalized regression models. Model ACC precision recall F1 MCC AUROC BRIER Ridge 0.889 0.852 0.958 0.902 0.782 0.964 0.129 Lasso 0.944 0.957 0.938 0.947 0.889 0.991 0.042 Elastic Net 0.978 0.979 0.979 0.979 0.955 0.999 0.023 EPSGO Elastic Net 0.989 1.000 0.979 0.989 0.978 1.000 0.018 AUROC, area under curve of receiver operating characteristic; ACC, accuracy; MCC, Matthews correlation coefficient; EPSGO, Efficient Parameter Selection via Global Optimization algorithm. -

Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in Vitro and in Vivo
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Genetik Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in vitro and in vivo - Towards Response Prediction in Cancer Therapy with Kinase Inhibitors Michaela Bairlein Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ. -Prof. Dr. K. Schneitz Prüfer der Dissertation: 1. Univ.-Prof. Dr. A. Gierl 2. Hon.-Prof. Dr. h.c. A. Ullrich (Eberhard-Karls-Universität Tübingen) 3. Univ.-Prof. A. Schnieke, Ph.D. Die Dissertation wurde am 07.01.2010 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 19.04.2010 angenommen. FOR MY PARENTS 1 Contents 2 Summary ................................................................................................................................................................... 5 3 Zusammenfassung .................................................................................................................................................... 6 4 Introduction .............................................................................................................................................................. 8 4.1 Cancer .............................................................................................................................................................. -

Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action
International Journal of Molecular Sciences Review Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action Paweł Łukasik 1, Irena Baranowska-Bosiacka 2, Katarzyna Kulczycka 3 and Izabela Gutowska 1,* 1 Department of Medical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 2 Department of Biochemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] 3 Department of Pharmaceutical Chemistry, Pomeranian Medical University in Szczecin, Powstancow Wlkp. 72 Av., 70-111 Szczecin, Poland; [email protected] * Correspondence: [email protected] Abstract: Recent studies on cyclin-dependent kinase (CDK) inhibitors have revealed that small molecule drugs have become very attractive for the treatment of cancer and neurodegenerative disorders. Most CDK inhibitors have been developed to target the ATP binding pocket. However, CDK kinases possess a very similar catalytic domain and three-dimensional structure. These features make it difficult to achieve required selectivity. Therefore, inhibitors which bind outside the ATP binding site present a great interest in the biomedical field, both from the fundamental point of view and for the wide range of their potential applications. This review tries to explain whether the ATP competitive inhibitors are still an option for future research, and highlights alternative approaches to discover more selective and potent small molecule inhibitors. Keywords: cyclin-dependent kinase inhibitors; cancer; cell cycle; CDKs; CDK inhibitors Citation: Łukasik, P.; Baranowska-Bosiacka, I.; Kulczycka, K.; Gutowska, I. Inhibitors of Cyclin-Dependent Kinases: Types 1. Cyclin-Dependent Kinases (CDKs) and Their Mechanism of Action. -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5. -
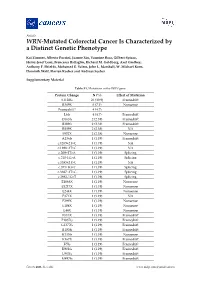
WRN-Mutated Colorectal Cancer Is Characterized by a Distinct Genetic Phenotype
Article WRN-Mutated Colorectal Cancer Is Characterized by a Distinct Genetic Phenotype Kai Zimmer, Alberto Puccini, Joanne Xiu, Yasmine Baca, Gilbert Spizzo, Heinz-Josef Lenz, Francesca Battaglin, Richard M. Goldberg, Axel Grothey, Anthony F. Shields, Mohamed E. Salem, John L. Marshall, W. Michael Korn, Dominik Wolf, Florian Kocher and Andreas Seeber Supplementary Material Table S1. Mutations in the WRN gene. Protein Change N (%) Effect of Mutation S1128fs 26 (30.9) Frameshift R369X 6 (7.1) Nonsense Frameshift* 4 (4.7) L6fs 4 (4.7) Frameshift D163fs 2 (2.38) Frameshift R389fs 2 (2.38) Frameshift R889X 2 (2.38) NA S952X 2 (2.38) Nonsense A136fs 1 (1.19) Frameshift c.1269+2T>C 1 (1.19) NA c.1898+2T>G 1 (1.19) NA c.209+2T>A 1 (1.19) Splicing c.210-1G>A 1 (1.19) Splicing c.3383+2T>C 1 (1.19) NA c.355+1G>T 1 (1.19) Splicing c.3687+2T>G 1 (1.19) Splicing c.3983-1G>T 1 (1.19) Splicing E1068X 1 (1.19) Nonsense E1217X 1 (1.19) Nonsense E244X 1 (1.19) Nonsense E371X 1 (1.19) NA E399X 1 (1.19) Nonsense E488X 1 (1.19) Nonsense E48X 1 (1.19) Nonsense E513X 1 (1.19) Frameshift F1037fs 1 (1.19) Frameshift G1377fs 1 (1.19) Frameshift I1183fs 1 (1.19) Frameshift K135fs 1 (1.19) Nonsense K167X 1 (1.19) Frameshift K5fs 1 (1.19) Frameshift K901fs 1 (1.19) Frameshift L967fs 1 (1.19) Frameshift M497fs 1 (1.19) Frameshift Cancers 2020, 12, x; doi: www.mdpi.com/journal/cancers Cancers 2020, 12 S2 of 13 N166fs 1 (1.19) Frameshift Q11fs 1 (1.19) Frameshift R1305X 1 (1.19) Nonsense R279fs 1 (1.19) Frameshift R565X 1 (1.19) Nonsene R741X 1 (1.19) Nonsense R987X 1 (1.19) NA T1011fs 1 (1.19) Frameshift W1014X 1 (1.19) Nonsense Y57X 1 (1.19) Nonsense Grand Total 84 *4 Frameshift mutations could not be described more precisely. -

Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation
REPORT Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation Javier A. Couto,1,6 August Y. Huang,2,6 Dennis J. Konczyk,1 Jeremy A. Goss,1 Steven J. Fishman,3 John B. Mulliken,1 Matthew L. Warman,2,4,5 and Arin K. Greene1,* Arteriovenous malformation (AVM) is a fast-flow, congenital vascular anomaly that may arise anywhere in the body. AVMs typically progress, causing destruction of surrounding tissue and, sometimes, cardiac overload. AVMs are difficult to control; they often re-expand after embolization or resection, and pharmacologic therapy is unavailable. We studied extracranial AVMs in order to identify their biological basis. We performed whole-exome sequencing (WES) and whole-genome sequencing (WGS) on AVM tissue from affected individuals. Endothelial cells were separated from non-endothelial cells by immune-affinity purification. We used droplet digital PCR (ddPCR) to confirm mutations found by WES and WGS, to determine whether mutant alleles were enriched in endothelial or non-endo- thelial cells, and to screen additional AVM specimens. In seven of ten specimens, WES and WGS detected and ddPCR confirmed somatic mutations in mitogen activated protein kinase kinase 1 (MAP2K1), the gene that encodes MAP-extracellular signal-regulated kinase 1 (MEK1). Mutant alleles were enriched in endothelial cells and were not present in blood or saliva. 9 of 15 additional AVM specimens contained mutant MAP2K1 alleles. Mutations were missense or small in-frame deletions that affect amino acid residues within or adja- cent to the protein’s negative regulatory domain. Several of these mutations have been found in cancers and shown to increase MEK1 activity. -

The Curing AI for Precision Medicine
The Curing AI for Precision Medicine Hoifung Poon 1 Medicine Today Is Imprecise Top 20 drugs 80% non-responders Wasted 1/3 health spending $750 billion / year 2 Disruption 1: Big Data 2009 2013: 40% 93% 3 Disruption 2: Pay-for-Performance Goal: 75% by 2020 4 Vemurafenib on BRAF-V600 Melanoma Before Treatment 15 Weeks 5 Vemurafenib on BRAF-V600 Melanoma Before Treatment 15 Weeks 23 Weeks 6 Why We Haven’t Solved Precision Medicine? … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … … ATTCGGATATTTAAGGC … … ATTCGGGTATTTAAGCC … High-Throughput Data Discovery Bottleneck #1: Knowledge Bottleneck #2: Reasoning AI is the key to overcome these bottlenecks 7 Use Case: Molecular Tumor Board 8 www.ucsf.edu/news/2014/11/120451/bridging-gap-precision-medicine Use Case: Molecular Tumor Board Problem: Hard to scale U.S. 2015: 1.6 million new cases, 600K deaths 902 cancer hospitals Memorial Sloan Kettering 2016: Sequence: Tens of thousand Board can review: A few hundred Wanted: Decision support for cancer precision medicine 9 First-Generation Molecular Tumor Board Knowledge bottleneck E.g., given a tumor sequence, determine: What genes and mutations are important What drugs might be applicable Can do manually but hard to scale 10 Next-Generation Molecular Tumor Board Reasoning bottleneck E.g., personalize drug combinations Can’t do manually, ever 11 Big Medical Data Decision Support Precision Medicine Machine Predict Reading Drug Combo 12 13 PubMed 26 millions abstracts Two new abstracts every minute Adds over one million every year 14 Machine Reading PMID: 123 … VDR+ binds to SMAD3 to form … PMID: 456 Knowledge … JUN expression Base is induced by SMAD3/4 … …… 15 Machine Reading Involvement of p70(S6)-kinase activation in IL-10 up-regulation in human monocytes by gp41 envelope protein of human immunodeficiency virus type 1 .. -
Oncomine Comprehensive Assay V3 Empower Your Oncology Research with Proven Ion Torrent Technology
Oncomine Comprehensive Assay v3 Empower your oncology research with proven Ion Torrent technology The Oncomine Comprehensive Assay v3 is available for use on the Ion GeneStudio S5 Systems with Ion Chef Instrument or for use on the Genexus System The Ion Torrent™ Oncomine™ Comprehensive Assay v3 • Detects relevant SNVs, CNVs, gene fusions, and is a member of the family of Ion Torrent™ Oncomine™ indels from 161 unique cancer driver genes in one assays for clinical cancer research. Oncomine assays streamlined workfl ow are multiple-biomarker assays based on next-generation • Optimized and verifi ed for the Ion Chef™ Instrument and sequencing (NGS). They have been adopted by leading Ion GeneStudio™ S5 Systems with the Ion 540™ Chip, cancer institutions around the world, have been used to or the new Ion Torrent™ Genexus™ System with the profi le thousands of samples in diff erent translational and Ion Torrent™ GX5™ Chip, both enabling full automation clinical research projects, and have consistently delivered including automated library prep reliable results. “The requirement of a lower DNA input for the Oncomine assay Oncomine Comprehensive Assay v3 is a signifi cant advantage when primary samples are becoming • Content is based on the latest advances in clinical increasingly limited.” oncology research and also enriched for targets known to be associated with (or drive) childhood cancers John Bartlett, PhD Director of Transformative Pathology Platform • Based on robust Ion AmpliSeq™ technology, this assay Ontario Institute for Cancer Research -

Mutually Exclusive Recurrent KRAS and MAP2K1 Mutations in Rosai
Modern Pathology (2017) 30, 1367–1377 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 1367 Mutually exclusive recurrent KRAS and MAP2K1 mutations in Rosai–Dorfman disease Sofia Garces1, L Jeffrey Medeiros1, Keyur P Patel1, Shaoying Li1, Sergio Pina-Oviedo1, Jingyi Li1, Juan C Garces2, Joseph D Khoury1 and C Cameron Yin1 1Department of Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, TX, USA and 2Instituto Oncológico Nacional Dr Juan Tanca Marengo, Guayaquil, Ecuador Rosai–Dorfman disease is a histiocytic disorder with a poorly defined pathogenesis. Recent molecular studies have revealed recurrent mutations involving genes in the MAPK/ERK pathway in Langerhans cell histiocytosis and Erdheim–Chester disease. However, cases of Rosai–Dorfman disease have rarely been assessed. We performed next-generation sequencing to assess 134 genes on 21 cases of Rosai–Dorfman disease, including 13 women and 8 men with a median age of 43 years (range, 3–82). In all, 13 had extranodal, 5 had nodal, and 3 had coexistent nodal and extranodal disease. The head and neck region was the most common area involved (n = 7). Mutation analysis detected point mutations in 7 (33%) cases, including KRAS (n = 4) and MAP2K1 (n = 3). No mutations were identified in ARAF, BRAF, PIK3CA, or any other genes assessed. Immunohistochemistry demonstrated p-ERK overexpression in 3 cases, all harboring MAP2K1 mutations. Patients carrying mutated genes were younger (median age, 10 vs 53 years, P = 0.0347) with more pediatric patients (4/7 vs 1/14, P = 0.0251). The presence of mutations correlated with location being more common in the head and neck region; 6/7 (86%) mutated vs 1/14 (7%) unmutated cases (P = 0.0009). -

Pediatric Langerhans Cell Histiocytosis: State of the Science and Future Directions
Pediatric Langerhans Cell Histiocytosis: State of the Science and Future Directions Nirav H. Thacker, MD, and Oussama Abla, MD Dr Thacker is a clinical fellow in Abstract: Langerhans cell histiocytosis (LCH) is an inflammatory hematology/oncology at the Hospital neoplasm of myeloid origin characterized by the presence of clas- for Sick Children. Dr Abla is an associ- sic CD1a+/CD207+ cells. An ongoing debate over the grouping ate professor in the Department of of LCH was finally settled in favor of neoplasm after the discovery Pediatrics at the University of Toronto and a staff hematologist/oncologist of the BRAF V600E mutation in 2010. The pathologic cells were at the Hospital for Sick Children in found to involve an almost universal activation of the MAPK/ERK Toronto, Ontario, Canada. pathway, with mutations identified in most kinases upstream of ERK (RAS/RAF/MEK). The clinical presentation of LCH is a mixed Corresponding author: bag, ranging from self-resolving localized disease to fulminant, fatal Oussama Abla, MD disseminated disease. The current standard of care for patients with Division of Haematology/Oncology Department of Paediatrics multisystem LCH, who have high relapse rates, continues to be The Hospital for Sick Children combination treatment with vinblastine and prednisone. Patients 555 University Avenue treated with BRAF and MEK inhibitors have shown a significant and Toronto, ON M5G 1X8 sustained response in early-phase trials. During the current decade, Canada researchers have described an extensive genomic landscape for Tel: 1 (416) 813-7879 LCH that has significantly enlarged our understanding of the biol- E-mail: [email protected] ogy and pathogenesis of this disease, especially neurodegenerative LCH.