Regional Polymorphism Assessment of Myanmar 'Sein Ta Lone' Mango
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Village Tract of Mandalay Region !
!. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. !. Myanmar Information Management Unit !. !. !. Village Tract of Mandalay Region !. !. !. !. 95° E 96° E Tigyaing !. !. !. / !. !. Inn Net Maing Daing Ta Gaung Taung Takaung Reserved Forest !. Reserved Forest Kyauk Aing Mabein !. !. !. !. Ma Gyi Kone Reserved !. Forest Thabeikkyin !. !. Reserved Forest !. Let Pan Kyunhla Kone !. Se Zin Kone !. Kyar Hnyat !. !. Kanbalu War Yon Kone !. !. !. Pauk Ta Pin Twin Nge Mongmit Kyauk Hpyu !. !. !. Kyauk Hpyar Yae Nyar U !. Kyauk Gyi Kyet Na !. Reserved Hpa Sa Bai Na Go Forest Bar Nat Li Shaw Kyauk Pon 23° N 23° Kyauk War N 23° Kyauk Gyi Li Shaw Ohn Dan Lel U !. Chaung Gyi !. Pein Pyit !. Kin Tha Dut !. Gway Pin Hmaw Kyauk Sin Sho !. Taze !. !. Than Lwin Taung Dun Taung Ah Shey Bawt Lone Gyi Pyaung Pyin !. Mogoke Kyauk Ka Paing Ka Thea Urban !. Hle Bee Shwe Ho Weik Win Ka Bar Nyaung Mogoke Ba Mun !. Pin Thabeikkyin Kyat Pyin !. War Yae Aye !. Hpyu Taung Hpyu Yaung Nyaung Nyaung Urban Htauk Kyauk Pin Ta Lone Pin Thar Tha Ohn Zone Laung Zin Pyay Lwe Ngin Monglon !. Ye-U Khin-U !. !. !. !. !. Reserved Forest Shwe Kyin !. !. Tabayin !. !. !. !. Shauk !. Pin Yoe Reserved !. Kyauk Myaung Nga Forest SAGAING !. Pyin Inn War Nat Taung Shwebo Yon !. Khu Lel Kone Mar Le REGION Singu Let Pan Hla !. Urban !. Koke Ko Singu Shwe Hlay Min !. Kyaung !. Seik Khet Thin Ngwe Taung MANDALAY Se Gyi !. Se Thei Nyaung Wun Taung Let Pan Kyar U Yin REGION Yae Taw Inn Kani Kone Thar !. !. Yar Shwe Pyi Wa Di Shwe Done !. Mya Sein Sin Htone Thay Gyi Shwe SHAN Budalin Hin Gon Taing Kha Tet !. Thar Nyaung Pin Chin Hpo Zee Pin Lel Wetlet Kyun Inn !. -

TRENDS in MANDALAY Photo Credits
Local Governance Mapping THE STATE OF LOCAL GOVERNANCE: TRENDS IN MANDALAY Photo credits Paul van Hoof Mithulina Chatterjee Myanmar Survey Research The views expressed in this publication are those of the author, and do not necessarily represent the views of UNDP. Local Governance Mapping THE STATE OF LOCAL GOVERNANCE: TRENDS IN MANDALAY UNDP MYANMAR Table of Contents Acknowledgements II Acronyms III Executive Summary 1 1. Introduction 11 2. Methodology 14 2.1 Objectives 15 2.2 Research tools 15 3. Introduction to Mandalay region and participating townships 18 3.1 Socio-economic context 20 3.2 Demographics 22 3.3 Historical context 23 3.4 Governance institutions 26 3.5 Introduction to the three townships participating in the mapping 33 4. Governance at the frontline: Participation in planning, responsiveness for local service provision and accountability 38 4.1 Recent developments in Mandalay region from a citizen’s perspective 39 4.1.1 Citizens views on improvements in their village tract or ward 39 4.1.2 Citizens views on challenges in their village tract or ward 40 4.1.3 Perceptions on safety and security in Mandalay Region 43 4.2 Development planning and citizen participation 46 4.2.1 Planning, implementation and monitoring of development fund projects 48 4.2.2 Participation of citizens in decision-making regarding the utilisation of the development funds 52 4.3 Access to services 58 4.3.1 Basic healthcare service 62 4.3.2 Primary education 74 4.3.3 Drinking water 83 4.4 Information, transparency and accountability 94 4.4.1 Aspects of institutional and social accountability 95 4.4.2 Transparency and access to information 102 4.4.3 Civil society’s role in enhancing transparency and accountability 106 5. -

Weekly Security Review (27 August – 2 September 2020)
Commercial-In-Confidence Weekly Security Review (27 August – 2 September 2020) Weekly Security Review Safety and Security Highlights for Clients Operating in Myanmar 27 August – 2 September 2020 Page 1 of 27 Commercial-In-Confidence Weekly Security Review (27 August – 2 September 2020) EXECUTIVE SUMMARY ............................................................................................................................. 3 Internal Conflict ....................................................................................................................................... 4 Nationwide .......................................................................................................................................... 4 Rakhine State ....................................................................................................................................... 4 Shan State ............................................................................................................................................ 5 Myanmar and the World ......................................................................................................................... 8 Election Watch ........................................................................................................................................ 8 Social and Political Stability ................................................................................................................... 11 Transportation ...................................................................................................................................... -

Health Facilities in Kyaukse Township - Mandalay
Myanmar Information Management Unit (! Health Facilities in Kyaukse Township - Mandalay 96°0'E 96°10'E 96°20'E 96°30'E 96°40'E PATHEINGYI 21°50'N 21°50'N AMARAPURA Kone Thar (190641) (Ohn Kyaw) Ü ! Ohn Kyaw (190639) (Ohn Kyaw) ! Za Yit Khe (190644) He Lel (190642) (Za Yit Khe) (Ohn Kyaw) ! ! Tha Man Thar (190638) ! (Tha Yet Pin) v® PYINOOLWIN Sintgaing (! SINTGAING Tha Yet Pin (190637) ! (Tha Yet Pin) v® v®! Ye (190636) (Ye) ! Shar Pin (190439) v® Kyaung (190645) (Shar Pin) ! (Kyaung) 21°40'N 21°40'N Let Pan (190474) Ngar Oe (190468) (Let Pan) (Ngar Oe) ! ! Kyaung (190472) Kyaung Kone (190473) Tha Pyay Wun (218235) ! Ywar Nan (190469) (Taung Nauk) (Taung Nauk) (Ye Baw Gyi) (Ywar Nan) ! ! ! Taung Hlwea (190491) Nyaung Pin Zauk (190455) Ngar Su (190441) Yae Twin Pyayt (190466) ! (Thin Taung) (Ngar Su) (Kyaung Pan Kone) ! Dway Hla (190440) ! ! (Thin Boke) ! (Dway Hla) ! ! v® ! Thin Taung (190490) v® Kyaung Pan Kone (190454) Taung Nauk (190470) (Thin Taung) ! Ah Shey Nge Toe (190495) Ywar Taw (190445) (Kyaung Pan Kone) (Taung Nauk) ! Ye Baw Lay (190499) Nyaung Shwe (190456) Ta Dar U Lay (190467) (Ah Shey Nge Toe) (Ye Baw Gyi) (Kyee Eik) Zay Kone (190465) ! v® Ye Baw Gyi (190498) ! (Nyaung Shwe) (Thin Boke) Shwe Dar (190496) (Ye Baw Gyi) ! ! (Thin Boke) ! Nyaung Wun (190457) Shwe Lay (190489) !(Shwe Dar) (Nyaung Wun) ! (In Daing) Kyee Eik (190443) ! Thin Pyo (190442) ! Thin Boke (190464) (Kyee Eik) Pa Daung Khar (190459) (Thin Pyo) (Thin Boke) KYAUKSE ! (Nyaung Wun) ! In Daing (190488) Thar Si (190497) ! ! Htan Zin Taw (190492) (Shwe Dar) -

Thu Zar Win October 2019
IMPACT OF SHWE HLAN BO AND HTEE TAW MOE RIVER PUMPING PROJECTS ON THE FARM HOUSEHOLDS OF BENEFICIAL AREA IN SINTGAING TOWNSHIP THU ZAR WIN OCTOBER 2019 IMPACT OF SHWE HLAN BO AND HTEE TAW MOE RIVER PUMPING PROJECTS ON THE FARM HOUSEHOLDS OF BENEFICIAL AREA IN SINTGAING TOWNSHIP THU ZAR WIN A thesis submitted to the post-graduate committee of the Yezin Agricultural University as a partial fulfillment of the requirements for the degree of Master of Agricultural Science (Agronomy) Department of Agronomy Yezin Agricultural University Nay Pyi Taw, Myanmar OCTOBER 2019 ii The thesis attached hereto, entitled “Impact of Shwe Hlan Bo and Htee Taw Moe River Pumping Projects on the Farm Households of Beneficial Area in Sintgaing Township” was prepared under the direction of the chairperson of the candidate supervisory committee and has been approved by all members of that committee and board of examiners as a partial fulfillment of the requirements for the degree of Master of Agricultural Science (Agronomy). ----------------------- ----------------------- Dr. Aye Aye Khaing Dr. Nyi Nyi Chairperson, Supervisory Committee External Examiner Associate Professor Assistant Director Department of Agronomy Agricultural Extension Division Yezin Agricultural University Department of Agriculture Yezin, Nay Pyi Taw Nay Pyi Taw ----------------------- ----------------------- Dr. Nyein Nyein Htwe Dr. Nay Myo Aung Member, Supervisory Committee Member, Supervisory Committee Professor and Head Director and Head Department of Agricultural Extension Department of Administration Yezin Agricultural University Yezin Agricultural University Yezin, Nay Pyi Taw Yezin, Nay Pyi Taw ----------------------- Dr. Hla Than Professor and Head Department of Agronomy Yezin Agricultural University Yezin, Nay Pyi Taw Date ------------------------ iii This thesis was submitted to the Rector of the Yezin Agricultural University and was accepted as a partial fulfillment of the requirements for the degree of Master of Agricultural Science (Agronomy). -
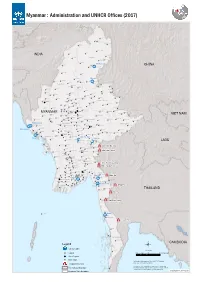
Myanmar : Administration and UNHCR Offices (2017)
Myanmar : Administration and UNHCR Offices (2017) Nawngmun Puta-O Machanbaw Khaunglanhpu Nanyun Sumprabum Lahe Tanai INDIA Tsawlaw Hkamti Kachin Chipwi Injangyang Hpakan Myitkyina Lay Shi Myitkyina CHINA Mogaung Waingmaw Homalin Mohnyin Banmauk Bhamo Paungbyin Bhamo Tamu Indaw Shwegu Momauk Pinlebu Katha Sagaing Mansi Muse Wuntho Konkyan Kawlin Tigyaing Namhkan Tonzang Mawlaik Laukkaing Mabein Kutkai Hopang Tedim Kyunhla Hseni Manton Kunlong Kale Kalewa Kanbalu Mongmit Namtu Taze Mogoke Namhsan Lashio Mongmao Falam Mingin Thabeikkyin Ye-U Khin-U Shan (North) ThantlangHakha Tabayin Hsipaw Namphan ShweboSingu Kyaukme Tangyan Kani Budalin Mongyai Wetlet Nawnghkio Ayadaw Gangaw Madaya Pangsang Chin Yinmabin Monywa Pyinoolwin Salingyi Matman Pale MyinmuNgazunSagaing Kyethi Monghsu Chaung-U Mongyang MYANMAR Myaung Tada-U Mongkhet Tilin Yesagyo Matupi Myaing Sintgaing Kyaukse Mongkaung VIET NAM Mongla Pauk MyingyanNatogyi Myittha Mindat Pakokku Mongping Paletwa Taungtha Shan (South) Laihka Kunhing Kengtung Kanpetlet Nyaung-U Saw Ywangan Lawksawk Mongyawng MahlaingWundwin Buthidaung Mandalay Seikphyu Pindaya Loilen Shan (East) Buthidaung Kyauktaw Chauk Kyaukpadaung MeiktilaThazi Taunggyi Hopong Nansang Monghpyak Maungdaw Kalaw Nyaungshwe Mrauk-U Salin Pyawbwe Maungdaw Mongnai Monghsat Sidoktaya Yamethin Tachileik Minbya Pwintbyu Magway Langkho Mongpan Mongton Natmauk Mawkmai Sittwe Magway Myothit Tatkon Pinlaung Hsihseng Ngape Minbu Taungdwingyi Rakhine Minhla Nay Pyi Taw Sittwe Ann Loikaw Sinbaungwe Pyinma!^na Nay Pyi Taw City Loikaw LAOS Lewe -

Study on Lotus Fiber Production in Sunn Ye Inn, Sintgaing Township
344 Banmaw University Research Journal 2020, Vol. 11, No.1 Study on Lotus Fiber Production in Sunn Ye Inn, Sintgaing Township San Win Abstract The lotus, Nelumbo nucifera Gaertn. is grown naturally in Sunn Ye Inn in Sintgaing Township, Mandalay Region. The lotus stem especially leaf petiole is being extentsively used in the extraction of lotus fibers. It can be weaved to make the famous of lotus robe (Kyathingan) and other cloth materials. The raw material maily lotus fibers are obtained from Inn Khone Village in Sunn Ye Inn. The Sunn Ye Inn is the sources of lotus fiber production and foods for people. The N.nucifera Gaertn. is influenced on ecological impact of Sunn Ye Inn and eco-social impact of local people. Key words: Lotus, Nelumbo nucifera Gaertn, ecological impact, eco-social impact. Introduction Lotus plant has symbol of religion and spiritual power in history of Myanmar. Buddhists considered lotus flower represents the success, peaceful, fresh, graceful and pure mind. Because of its pristine value, Padonma Kya is also known as the sacred lotus. Ancient Myanmar Kings used lotus flower, bud, stems and petals in wall painting and statue (SK Myanmar Cultural Heritage 2016). All parts of lotus plant are useful, stem for producing lotus fiber, flower for offering to Buddha, leaves for packing food, meat etc. Lotus leaves are also used as plate to decorate food. Lotus root and seed are also edible (Hlaing 2016). Nelumbo nucifera Gaertn. is an aquatic perennial belonging to the family of Nelumbonaceae, which has several common names (e.g. Indian lotus, Chinese water lily, and sacred lotus), lotus is a large and rhizomatous aquatic herb with slender, elongated, branched, creeping stem consisting of nodal roots; leaves are large, peltate; petioles are long, rough with small distinct prickles; flowers are white to rosy, sweet-scented, solitary, hermaphrodite, 10-25 cm diameter; ripe carpel are 12mm long, ovoid and glabrous; fruits are ovoid having nut like achene; seeds are black, hard and ovoid (Sridhar and Rajeev 2007).The lotus, N. -

Mandalay, Pathein and Mawlamyine - Mandalay, Pathein and Mawlamyine
Urban Development Plan Development Urban The Republic of the Union of Myanmar Ministry of Construction for Regional Cities The Republic of the Union of Myanmar Urban Development Plan for Regional Cities - Mawlamyine and Pathein Mandalay, - Mandalay, Pathein and Mawlamyine - - - REPORT FINAL Data Collection Survey on Urban Development Planning for Regional Cities FINAL REPORT <SUMMARY> August 2016 SUMMARY JICA Study Team: Nippon Koei Co., Ltd. Nine Steps Corporation International Development Center of Japan Inc. 2016 August JICA 1R JR 16-048 Location業務対象地域 Map Pannandin 凡例Legend / Legend � Nawngmun 州都The Capital / Regional City Capitalof Region/State Puta-O Pansaung Machanbaw � その他都市Other City and / O therTown Town Khaunglanhpu Nanyun Don Hee 道路Road / Road � Shin Bway Yang � 海岸線Coast Line / Coast Line Sumprabum Tanai Lahe タウンシップ境Township Bou nd/ Townshipary Boundary Tsawlaw Hkamti ディストリクト境District Boundary / District Boundary INDIA Htan Par Kway � Kachinhin Chipwi Injangyang 管区境Region/S / Statetate/Regi Boundaryon Boundary Hpakan Pang War Kamaing � 国境International / International Boundary Boundary Lay Shi � Myitkyina Sadung Kan Paik Ti � � Mogaung WaingmawミッチMyitkyina� ーナ Mo Paing Lut � Hopin � Homalin Mohnyin Sinbo � Shwe Pyi Aye � Dawthponeyan � CHINA Myothit � Myo Hla Banmauk � BANGLADESH Paungbyin Bhamo Tamu Indaw Shwegu Katha Momauk Lwegel � Pinlebu Monekoe Maw Hteik Mansi � � Muse�Pang Hseng (Kyu Koke) Cikha Wuntho �Manhlyoe (Manhero) � Namhkan Konkyan Kawlin Khampat Tigyaing � Laukkaing Mawlaik Tonzang Tarmoenye Takaung � Mabein -

Myanmar Situation Update (12 - 18 July 2021)
Myanmar Situation Update (12 - 18 July 2021) Summary Myanmar detained State Counsellor Daw Aung San Suu Kyi, President U Win Myint and former Naypyitaw Council Chairman Dr. Myo Aung appeared at a special court in Naypyitaw’s Zabuthiri township for their trial for incitement under Section 505(b) of the Penal Code. The junta filed fresh charges against Suu Kyi, bringing the number of cases she faces to ten with a potential prison sentence of 75 years. The next court hearings of their trial have been moved to July 26 and 27, following the junta’s designation of a week-long public holiday and lockdown order. Senior National League for Democracy (NLD) patron Win Htein was indicted on a sedition charge by a court inside a Naypyitaw detention center with a possible prison sentence of up to 20 years. The state- run MRTV also reported the Anti-Corruption Commission (AAC) made a complaint against the former Chief Minister of Shan State, three former state ministers, and three people under the anti-corruption law at Taunggyi Township police station while the junta has already filed corruption cases against many former State Chief Ministers under the NLD government. During the press conference on 12 July 2021 in Naypyitaw, the junta-appointed Union Election Commission (UEC) announced that 11,305,390 voter list errors were found from the investigation conducted by the UEC. The UEC also said the Ministry of Foreign Affairs is investigating foreign funding of political parties and the investigation reports will be published soon with legal actions to be taken against the parties who violate the law. -

Eligible Voters Per Pyithu Hluttaw Constituency 2015 Elections
Myanmar Information Management Unit Eligible Voters per Pyithu Hluttaw Constituency 2015 Elections 90° E 95° E 100° E This map shows the variation in the number of registered voters per township according to UEC data. Nawngmun BHUTAN Puta-O Machanbaw Nanyun Khaunglanhpu Sumprabum Tsawlaw Tanai Lahe Injangyang INDIA Hpakant KACHIN Hkamti Chipwi Hpakant Waingmaw Lay Shi Mogaung N N ° CHINA ° 5 Homalin Myitkyina 5 2 Mohnyin 2 Momauk Banmauk Indaw BANGLADESH Shwegu Bhamo PaungbySinAGAING Katha Tamu Pinlebu Konkyan Wuntho Mansi Muse Kawlin Tigyaing Tonzang Mawlaik Namhkan Kutkai Laukkaing Mabein Kyunhla Thabeikkyin Kunlong Tedim Manton Hopang Kalewa Hseni Kale Kanbalu Mongmit Taze Namtu Hopang Falam Namhsan Lashio Mongmao Mingin Ye-U Mogoke Pangwaun Thantlang Khin-U Tabayin Kyaukme Shwebo Singu Tangyan Narphan Kani Hakha Budalin Wetlet Nawnghkio Mongyai Pangsang Ayadaw Madaya Hsipaw Yinmabin Monywa Sagaing Patheingyi Gangaw Salingyi VIETNAM Pale Myinmu Mongyang Matupi Chaung-U Ngazun Pyinoolwin Kyethi Myaung Matman CHIN Tilin Myaing Sintgaing Mongkaung Monghsu Mongkhet Tada-U Kyaukse Lawksawk Mongla Pauk Myingyan Paletwa Mindat Yesagyo Natogyi Saw Myittha SHAN Pakokku Hopong Laihka Maungdaw Ywangan Kunhing Mongping Kengtung Mongyawng MTaAunNgthDa ALWAundYwin Buthidaung Kanpetlet Seikphyu Nyaung-U Mahlaing Pindaya Loilen Kyauktaw Nansang Monghpyak Kyaukpadaung Meiktila Thazi Taunggyi Chauk Salin Kalaw Mongnai Ponnagyun Pyawbwe Tachileik Minbya Monghsat Rathedaung Mrauk-U Sidoktaya Yenangyaung Nyaungshwe RAKHINE Natmauk Yamethin Pwintbyu Mawkmai -

Dry Zone and South East Region - Myanmar
Myanmar Information Management Unit Dry Zone and South East Region - Myanmar !( !( !( !( Manhlyoe Muse (Manhero) !( !( Cikha Wuntho !( !( Namhkan Konkyan !( !( Khampat Kawlin !( !( Tigyaing !( Laukkaing !( Mawlaik Tonzang !( !( Tarmoenye !( BHUTAN Takaung !( Mabein Chinshwehaw Namtit Kutkai !( !( !( Kachin !( Hopang INDIA Kunlong!( State Tedim !( Rihkhawdar !( !( Kyunhla Hseni !( !( CHINA Manton Pan Lon !( !( Sagaing Kale Kalewa Kanbalu Region !( !( !( Mongmit !( Namtu Ü Taze !( Kanbalu Pangwaun INDIA !( Namhsan Mongmao Chin Shan Taze Lashio !( !( State State Falam !( Mogoke !( Mandalay !( Mingin Thabeikkyin !( Region !( !( Ye-U Rakhine Magway Monglon State Ye-U Khin-U !( Mongngawt Region !( !( Khin-U !( CHINA LAOS Thantlang Tabayin Man Kan Kayah !( Hakha !( !( State Tabayin Kyauk Hsipaw Namphan Bago .! Myaung !( Shwebo !( !( Region SAGAING Shwebo Singu !( !( Kyaukme REGION !( Tangyan !( THAILAND Ayeyarwady Yangon Kayin Kani Mongyai State !( Budalin !( Region Region !( Budalin Wetlet Ayadaw !( Nawnghkio !( Wein Ayadaw !( !( Wetlet Mon State Madaya Gangaw !( Pangsang !( !( Monywa Yinmabin Tanintharyi !( Monywa Rezua !( Yinmabin Mandalay Region !( Sagaing City Pyinoolwin Mongpauk Salingyi Myinmu !( !( Pale !( Chaung-U .! Matman Pale !( Myinmu Kyethi !( !( Monghsu Chaung-U !( Ngazun Sagaing !( Salingyi !( !( BANGLADESH Myaung ! Myitnge Mongyang . !( !( !( !( Tada-U Ngazun CHIN Monghsu Mongkhet Myaung Sintgaing !( STATE Tilin Tada-U !( Mongkaing Kyethi Mongsan Mongla !( (Hmonesan) Mongnawng !( Myaing Yesagyo Intaw !( !( Matupi Kyaukse Kyaukse -
Administrative Map
Myanmar Information Management Unit Myanmar Administrative Map 94°E 96°E 98°E 100°E India China Bhutan Bangladesh Along India Vietnam KACHIN Myanmar Dong Laos South China Sea Bay of Bengal / Passighat China Thailand Daporija Masheng SAGAING 28°N Andaman Sea Philippines Tezu 28°N Cambodia Sea of the Philippine Gulf of Thailand Bangladesh Pannandin !( Gongshan CHIN NAWNGMUN Sulu Sea Namsai Township SHAN MANDALAY Brunei Malaysia Nawngmun MAGWAY Laos Tinsukia !( Dibrugarh NAY PYI TAW India Ocean RAKHINE Singapore Digboi Lamadi KAYAH o Taipi Duidam (! !( Machanbaw BAGO Margherita Puta-O !( Bomdi La !( PaPannssaauunngg North Lakhimpur KHAUNGLANHPU Weixi Bay of Bengal Township Itanagar PUTA-O MACHANBAW Indonesia Township Township Thailand YAN GON KAY IN r Khaunglanhpu e !( AYE YARWADY MON v Khonsa i Nanyun R Timor Sea (! Gulf of Sibsagar a Martaban k Fugong H i l NANYUN a Township Don Hee M !( Jorhat Mon Andaman Sea !(Shin Bway Yang r Tezpur e TANAI v i TANINTHARYI NNaaggaa Township R Sumprabum !( a Golaghat k SSeellff--AAddmmiinniisstteerreedd ZZoonnee SUMPRABUM Township i H Gulf of a m Thailand Myanmar administrative Structure N Bejiang Mangaldai TSAWLAW LAHE !( Tanai Township Union Territory (1) Nawgong(nagaon) Township (! Lahe State (7) Mokokchung Tuensang Lanping Region (7) KACHIN INDIA !(Tsawlaw Zunheboto Hkamti INJANGYANG Hojai Htan Par Kway (! Township !( 26°N o(! 26°N Dimapur !( Chipwi CHIPWI Liuku r Township e Injangyang iv !( R HKAMTI in w Township d HPAKANT MYITKYINA Lumding n i Township Township Kohima Mehuri Ch Pang War !(Hpakant