SSR Based Characterization of Indigenous Harnai Sheep Breed of Balochistan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Section Iv District Profiles Awaran
SECTION IV DISTRICT PROFILES AWARAN Awaran district lies in the south of the Balochistan province. Awaran is known as oasis of AGRICULTURAL INFORMATION dates. The climate is that of a desert with hot summer and mild winter. Major crops include Total cultivated area (hectares) 23,600 wheat, barley, cotton, pulses, vegetable, fodder and fruit crops. There are three tehsils in the district: Awaran, Jhal Jhao and Mashkai. The district headquarter is located at Awaran. Total non-cultivated area (hectares) 187,700 Total area under irrigation (hectares) 22,725 Major rabi crop(s) Wheat, vegetable crops SOIL ATTRIBUTES Mostly barren rocks with shallow unstable soils Major kharif crop(s) Cotton, sorghum Soil type/parent material material followed by nearly level to sloppy, moderately deep, strongly calcareous, medium Total livestock population 612,006 textured soils overlying gravels Source: Crop Reporting Services, Balochistan; Agriculture Census 2010; Livestock Census 2006 Dominant soil series Gacheri, Khamara, Winder *pH Data not available *Electrical conductivity (dS m-1) Data not available Organic matter (%) Data not available Available phosphorus (ppm) Data not available Extractable potassium (ppm) Data not available Farmers availing soil testing facility (%) 2 (Based on crop production zone wise data) Farmers availing water testing facility (%) 0 (Based on crop production zone wise data) Source: District Soil Survey Reports, Soil Survey of Pakistan Farm Advisory Centers, Fauji Fertilizer Company Limited (FFC) Inputs Use Assessment, FAO (2018) Land Cover Atlas of Balochistan (FAO, SUPARCO and Government of Balochistan) Source: Information Management Unit, FAO Pakistan *Soil pH and electrical conductivity were measured in 1:2.5, soil:water extract. -

Pashto, Waneci, Ormuri. Sociolinguistic Survey of Northern
SOCIOLINGUISTIC SURVEY OF NORTHERN PAKISTAN VOLUME 4 PASHTO, WANECI, ORMURI Sociolinguistic Survey of Northern Pakistan Volume 1 Languages of Kohistan Volume 2 Languages of Northern Areas Volume 3 Hindko and Gujari Volume 4 Pashto, Waneci, Ormuri Volume 5 Languages of Chitral Series Editor Clare F. O’Leary, Ph.D. Sociolinguistic Survey of Northern Pakistan Volume 4 Pashto Waneci Ormuri Daniel G. Hallberg National Institute of Summer Institute Pakistani Studies of Quaid-i-Azam University Linguistics Copyright © 1992 NIPS and SIL Published by National Institute of Pakistan Studies, Quaid-i-Azam University, Islamabad, Pakistan and Summer Institute of Linguistics, West Eurasia Office Horsleys Green, High Wycombe, BUCKS HP14 3XL United Kingdom First published 1992 Reprinted 2004 ISBN 969-8023-14-3 Price, this volume: Rs.300/- Price, 5-volume set: Rs.1500/- To obtain copies of these volumes within Pakistan, contact: National Institute of Pakistan Studies Quaid-i-Azam University, Islamabad, Pakistan Phone: 92-51-2230791 Fax: 92-51-2230960 To obtain copies of these volumes outside of Pakistan, contact: International Academic Bookstore 7500 West Camp Wisdom Road Dallas, TX 75236, USA Phone: 1-972-708-7404 Fax: 1-972-708-7433 Internet: http://www.sil.org Email: [email protected] REFORMATTING FOR REPRINT BY R. CANDLIN. CONTENTS Preface.............................................................................................................vii Maps................................................................................................................ -

Public Sector Development Programme 2019-20 (Original)
GOVERNMENT OF BALOCHISTAN PLANNING & DEVELOPMENT DEPARTMENT PUBLIC SECTOR DEVELOPMENT PROGRAMME 2019-20 (ORIGINAL) Table of Contents S.No. Sector Page No. 1. Agriculture……………………………………………………………………… 2 2. Livestock………………………………………………………………………… 8 3. Forestry………………………………………………………………………….. 11 4. Fisheries…………………………………………………………………………. 13 5. Food……………………………………………………………………………….. 15 6. Population welfare………………………………………………………….. 16 7. Industries………………………………………………………………………... 18 8. Minerals………………………………………………………………………….. 21 9. Manpower………………………………………………………………………. 23 10. Sports……………………………………………………………………………… 25 11. Culture……………………………………………………………………………. 30 12. Tourism…………………………………………………………………………... 33 13. PP&H………………………………………………………………………………. 36 14. Communication………………………………………………………………. 46 15. Water……………………………………………………………………………… 86 16. Information Technology…………………………………………………... 105 17. Education. ………………………………………………………………………. 107 18. Health……………………………………………………………………………... 133 19. Public Health Engineering……………………………………………….. 144 20. Social Welfare…………………………………………………………………. 183 21. Environment…………………………………………………………………… 188 22. Local Government ………………………………………………………….. 189 23. Women Development……………………………………………………… 198 24. Urban Planning and Development……………………………………. 200 25. Power…………………………………………………………………………….. 206 26. Other Schemes………………………………………………………………… 212 27. List of Schemes to be reassessed for Socio-Economic Viability 2-32 PREFACE Agro-pastoral economy of Balochistan, periodically affected by spells of droughts, has shrunk livelihood opportunities. -

Buffer Zone, Colonial Enclave, Or Urban Hub?
Working Paper no. 69 - Cities and Fragile States - BUFFER ZONE, COLONIAL ENCLAVE OR URBAN HUB? QUETTA :BETWEEN FOUR REGIONS AND TWO WARS Haris Gazdar, Sobia Ahmad Kaker, Irfan Khan Collective for Social Science Research February 2010 Crisis States Working Papers Series No.2 ISSN 1749-1797 (print) ISSN 1749-1800 (online) Copyright © H. Gazdar, S. Ahmad Kaker, I. Khan, 2010 24 Crisis States Working Paper Buffer Zone, Colonial Enclave or Urban Hub? Quetta: Between Four Regions and Two Wars Haris Gazdar, Sobia Ahmad Kaker and Irfan Khan Collective for Social Science Research, Karachi, Pakistan Quetta is a city with many identities. It is the provincial capital and the main urban centre of Balochistan, the largest but least populous of Pakistan’s four provinces. Since around 2003, Balochistan’s uneasy relationship with the federal state has been manifested in the form of an insurgency in the ethnic Baloch areas of the province. Within Balochistan, Quetta is the main shared space as well as a point of rivalry between the two dominant ethnic groups of the province: the Baloch and the Pashtun.1 Quite separately from the internal politics of Balochistan, Quetta has acquired global significance as an alleged logistic base for both sides in the war in Afghanistan. This paper seeks to examine different facets of Quetta – buffer zone, colonial enclave and urban hub − in order to understand the city’s significance for state building in Pakistan. State-building policy literature defines well functioning states as those that provide security for their citizens, protect property rights and provide public goods. States are also instruments of repression and the state-building process is often wrought with conflict and the violent suppression of rival ethnic and religious identities, and the imposition of extractive economic arrangements (Jones and Chandaran 2008). -

Balochistan Province Report on Mouza Census 2008
TABLE 1 NUMBER OF KANUNGO CIRCLES,PATWAR CIRCLES AND MOUZAS WITH STATUS NUMBER OF NUMBER OF MOUZAS KANUNGO CIRCLES/ PATWAR ADMINISTRATIVE UNIT PARTLY UN- SUPER- CIRCLES/ TOTAL RURAL URBAN FOREST URBAN POPULATED VISORY TAPAS TAPAS 1 2 3 4 5 6 7 8 9 BALOCHISTAN 179 381 7480 6338 127 90 30 895 QUETTA DISTRICT 5 12 65 38 15 10 1 1 QUETTA CITY TEHSIL 2 6 23 7 9 7 - - QUETTA SADDAR TEHSIL 2 5 38 27 6 3 1 1 PANJPAI TEHSIL 1 1 4 4 - - - - PISHIN DISTRICT 6 17 392 340 10 3 8 31 PISHIN TEHSIL 3 6 47 39 2 1 - 5 KAREZAT TEHSIL 1 3 39 37 - 1 - 1 HURAM ZAI TEHSIL 1 4 16 15 - 1 - - BARSHORE TEHSIL 1 4 290 249 8 - 8 25 KILLA ABDULLAH DISTRICT 4 10 102 95 2 2 - 3 GULISTAN TEHSIL 1 2 10 8 - - - 2 KILLA ABDULLAH TEHSIL 1 3 13 12 1 - - - CHAMAN TEHSIL 1 2 31 28 1 2 - - DOBANDI SUB-TEHSIL 1 3 48 47 - - - 1 NUSHKI DISTRICT 2 3 45 31 1 5 - 8 NUSHKI TEHSIL 1 2 26 20 1 5 - - DAK SUB-TEHSIL 1 1 19 11 - - - 8 CHAGAI DISTRICT 4 6 48 41 1 4 - 2 DALBANDIN TEHSIL 1 3 30 25 1 3 - 1 NOKUNDI TEHSIL 1 1 6 5 - - - 1 TAFTAN TEHSIL 1 1 2 1 - 1 - - CHAGAI SUB-TEHSIL 1 1 10 10 - - - - SIBI DISTRICT 6 15 161 124 7 1 6 23 SIBI TEHSIL 2 5 35 31 1 - - 3 KUTMANDAI SUB-TEHSIL 1 2 8 8 - - - - SANGAN SUB-TEHSIL 1 2 3 3 - - - - LEHRI TEHSIL 2 6 115 82 6 1 6 20 HARNAI DISTRICT 3 5 95 81 3 3 - 8 HARNAI TEHSIL 1 3 64 55 1 1 - 7 SHARIGH TEHSIL 1 1 16 12 2 1 - 1 KHOAST SUB-TEHSIL 1 1 15 14 - 1 - - KOHLU DISTRICT 6 18 198 195 3 - - - KOHLU TEHSIL 1 2 37 35 2 - - - MEWAND TEHSIL 1 5 38 37 1 - - - KAHAN TEHSIL 4 11 123 123 - - - - DERA BUGTI DISTRICT 9 17 224 215 4 1 - 4 DERA BUGTI TEHSIL 1 -

Frequency of Various Human Malaria Infections in Hottest Areas of Central Balochistan, Pakistan: Duki, Harnai, and Sibi
Various Human Malaria Infections in Central Balochistan Pak Armed Forces Med J 2008; 58(3): 276-285 FREQUENCY OF VARIOUS HUMAN MALARIA INFECTIONS IN HOTTEST AREAS OF CENTRAL BALOCHISTAN, PAKISTAN: DUKI, HARNAI, AND SIBI Mohammad Iqbal Yasinzai, Juma Khan Kakarsulemankhel University of Balochistan, Quetta ABSTRACT Objective: This study was designed to assess the frequency of malaria infection in human population Duki, Harnai and Sibi the hottest areas of centre Balochistan, Pakistan. Study Design: A descriptive study. Place and duration of study: The study was conducted in Balochistan from July 2004 to June 2006. Patients and Methods: This study was conducted in Duki (8 localities), Harnai (7 localities) and Sibi (10 localities) and malarial parasites were identified in the blood slides of suspected patients from July, 2004 to June, 2006 and encompassed 6730 subjects. Results: Out of 6730 suspected cases of malaria, 2304 (34.2%) were found to be positive for malarial parasite in blood smear slides. Out of positive cases, 1316 (57.1%) were identified as Plasmodium falciparum infection, 988 (42.8%) cases with P. vivax. Seasonal variation was also noted in Duki area with highest (81.8%:36/44) infection of P. falciparum. in April and lowest (72.2%:26/36) infection in December. In Harnai area highest (84.2%:32/38) infection of P. falciparum in December and lowest (61.1%:11/18) infection in January. Similarly, in Sibi area highest (81.5%:31/38) infection of P. vivax in August and lowest (58.8%:30/51) infection in May. Infection with P. falciparum in male in Duki area was 62.5 % (10/16), 68.9% (60/87) in Harnai, 63.1% (24/38), infection with P. -

Lithofacies, Depositional Environments, and Regional Stratigraphy of the Lower Eocene Ghazij Formation, Balochistan, Pakistan
U.S. Department of the Interior U.S. Geological Survey Lithofacies, Depositional Environments, and Regional Stratigraphy of the Lower Eocene Ghazij Formation, Balochistan, Pakistan U.S. Geological Survey Professional Paper 1599 Prepared in cooperation with the Geological Survey of Pakistan A Cover. Exposures of the lower Eocene Ghazij Formation along the northeast flank of the Sor Range, Balochistan, Pakistan. Photograph by Stephen B. Roberts. Lithofacies, Depositional Environments, and Regional Stratigraphy of the Lower Eocene Ghazij Formation, Balochistan, Pakistan By Edward A. Johnson, Peter D. Warwick, Stephen B. Roberts, and Intizar H. Khan U.S. GEOLOGICAL SURVEY PROFESSIONAL PAPER 1599 Prepared in cooperation with the Geological Survey of Pakistan UNITED STATES GOVERNMENT PRINTING OFFICE, WASHINGTON : 1999 U.S. DEPARTMENT OF THE INTERIOR BRUCE BABBITT, Secretary U.S. GEOLOGICAL SURVEY Charles G. Groat, Director For sale by U.S. Geological Survey, Information Services Box 25286, Federal Center Denver, CO 80225 Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. Government Library of Congress Cataloging-in-Publication Data Lithofacies, depositional environments, and regional stratigraphy of the lower Eocene Ghazij Formation, Balochistan, Pakistan / by Edward A. Johnson . .[et al.]. p. cm.—(U.S. Geological Survey professional paper ; 1599) "Prepared in cooperation with the Geological Survey of Pakistan." Includes bibliographical references. 1. Geology, Stratigraphic—Eocene. 2. Geology—Pakistan— Balochistan. 3. Coal—Geology—Pakistan—Balochistan. 4. Ghazij Formation (Pakistan). I. Johnson, Edward A. (Edward Allison), 1940- . II. Series. QE692.2.L58 1999 553.2'4'0954915—dc21 98-3305 ISBN=0-607-89365-6 CIP CONTENTS Abstract.......................................................................................................................... -

Abstract Insurgency Is a Violent Political Struggle Borne out of Grievances in a Local Community
Citation: Jilani, S. G., & Mujaddid, G. (2020). Theory and Practice of Insurgency and Counterinsurgency: The Case Study of Balochistan. Global Security and Strategic Studies Review, V(I), 1-13. https://doi.org/10.31703/gsssr.2020(V-I).01 Theory and Practice of Insurgency and Counterinsurgency: The Case Study of Balochistan Pages: 1 – 13 | Vol. V, No. I (Winter 2020) Sheikh Ghulam Jilani* | Ghulam Mujaddid† Abstract Insurgency is a violent political struggle borne out of grievances in a local community. The phenomenon of insurgency is old, and the ways of tackling insurgencies have always been studied with interest. Pakistan has faced sporadic insurgencies in its Balochistan province. This study applies conceptual framework propounded by Scott Moore to Balochistan insurgency and analyzes the complex combination of Actions triggered by varied Beliefs and Structures of the Baloch society. In order to employ effective countermeasures to insurgency, an integrated approach needs to be worked out that seeks to create positive changes in the intertwined dimensions of Actions, Beliefs and Structures. The study finds that such an approach to handle insurgency in Balochistan has generally been successful. And the Baloch society has largely been mainstreamed in the national society and the state of Pakistan. Key Words: Insurgency, Counterinsurgency, Balochistan, Pakistan, Actions, Beliefs and Structures, Scott Moore Introduction Insurgency is a politico-military struggle, carried through guerrilla warfare. As a form of political violence, the phenomenon of insurgency is as old as humanity itself. Guerrillas and partisans have existed throughout the recorded history, but their irregular conflict format has been transformed by the profound changes occurring in 20th century, when ‘social, economic, psychological, and, especially, political elements were combined with guerrilla tactics in order to radically alter the structure of the state by force’. -

The Multi-Layered Minority: Exploring the Intersection of Gender, Class and Religious-Ethnic Affiliation in the Marginalisation of Hazara Women in Pakistan
CREID INTERSECTIONS SERIES Religious Inequalities and Gender The Multi-Layered Minority: Exploring the Intersection of Gender, Class and Religious-Ethnic Affiliation in the Marginalisation of Hazara Women in Pakistan Sadiqa Sultan, Maryam Kanwer and Jaffer Abbas Mirza December 2020 Part of the CREID Intersection Series Collection on Violence and Discrimination Against Women of Religious Minority Backgrounds in Pakistan About CREID The Coalition for Religious Equality and Inclusive Development (CREID) provides research evidence and delivers practical programmes which aim to redress poverty, hardship, and exclusion resulting from discrimination on the grounds of religion or belief. CREID is an international consortium led by the Institute of Development Studies (IDS) and funded by UK aid from the UK Government. Key partners include Al-Khoei Foundation, Minority Rights Group (MRG), and Refcemi. Find out more: www.ids.ac.uk/creid. © Institute of Development Studies 2020 Front cover image credit: Surian Soosay CC BY-2.0 ISBN: 978-1-78118-728-9 DOI: 10.19088/CREID.2020.005 This is an Open Access paper distributed under the terms of the Creative Commons Attribution 4.0 International licence (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited and any modifications or adaptations are indicated. Available from: Coalition for Religious Equality and Inclusive Development (CREID), Institute of Development Studies (IDS), Brighton BN1 9RE, UK Tel: +44(0) 1273 915704 -

Balochis of Pakistan: on the Margins of History
BALOCHIS OF PAKISTAN: ON THE MARGINS OF HISTORY November 2006 First published in 2006 by The Foreign Policy Centre 23-28 Penn Street London N1 5DL www.fpc.org.uk Email: [email protected] © Foreign Policy Centre 2006 All rights reserved ISBN-13: 978-1-905833-08-5 ISBN-10: 1-905833-08-3 PREFACE The Foreign Policy Centre is keen to promote debate about some of the worlds lesser known conflicts. The situation in Balochistan is one such example. This pamphlet sets out a powerful and well argued case that the Balochi people have been let down - by the British Empire, by the founders of modern India and by successive Governments in Pakistan. It is a fascinating analysis which we hope will contribute to constructive discussion about Balochistans future. The Foreign Policy Centre Disclaimer : The views in this paper are not necessarily those of the Foreign Policy Centre. CONTENTS Baloch and Balochistan through History A Brief Prologue The Khanate of Kalat: Between Dependency and Sovereignty The Colonial Era: The British Policy of Divide et Empera Boundary Demarcation and Trifurcation of Baloch Terrain Pakistan absorbs the Khanate Partition and the Annexation of Balochistan The Indian Position Baloch Insurgencies 1948-1977 First Guerrilla Revolt The Second Revolt Third Balochi Resistance: The 1970s The State of Nationalist Politics Today Signifiers of Balochi Nationalism a) Language b) Islam c) Sardari System d) Aversion towards Punjabi and Pathan Immigration The Post-1980 Phase The Contemporary Socio-Political Scenario in Balochistan Influence of Jihad in Afghanistan Does Islam blunt Baloch nationalism? The Baloch Resistance Movement 2000-2006 The state of Baloch Insurgency Human Rights Violations Killing of Nawab Bugti Causes of Baloch Disaffection a) Richest in Resources, Yet the Poorest Province b) Lack of Representation c) The case for Autonomy d) Development as Colonisation The Future The Weaknesses The Road Ahead Endnotes ABSTRACT The Balochis, like the Kurds, their cousins from Aleppo, do not have a sovereign state of their own. -

Village List of Baluchistan , Pakistan
M. INT. 11-12. !511. CENSUS OF PAKISTAN, 1951 VILLAGE LIST. BALUCHISTAN DISTRICTS & STATES. 315.491 1952 195~ Sal Vii L Price 3181- ROVINClAL SUPERINTENDENT OF CE~SUS lN BAl,UCHISTAN, QUETTA. VILLAGE LIST OF BALl'CHISTAN. FOHEWOI:ll. This Village List is prepared from the data collected at the First Census of Pakistan during February 1951. It glV3S population of Towns and Villages, and preserves information which does not appear in full detail in the Census Tables. Part T of this vo"lume (pages 1-92) contains infnrmation pertaining to the Baluchistan Districts. P'ut II (pages 93-203) relates to the States Union. The source of area figures for Distrids and Tehsils, and States and Sub-Divisions, is Survey of Pakistan. Figures of area for smaller units are Hot available Figures of population shmvn again.:;t each Village and Town have been rounded to the nearest 10 Actu::tl numbers have been shown in the column i. Houses ". Similarly S11mmary Tables appearing in the begining of the village li~t for each District and State show actual Census figures. The local details are based on information furnished by Distri!. and State authorities. The following symbols have been u~ed :- Seh means Primary School. M. Sch. " ~ljddlc School. H. Seh. " High School. G. Sch. " Girls Sell 001 P. O. " Post Office. T. O. "Telegraph Office. P. T. O. ., Post and Telegraph Office. P. S. Police Station. " L P. Levy Post. " Ry. ~, Railway Station. Hasp. ,, Hospital. D. B. Dak Bungalow. " R.H. Rest House. " PART I. BALUCHISTAN DISTRICTS. CONTENTS Page Chagai 1 Loralai 9 Quetta-Pishin .. -
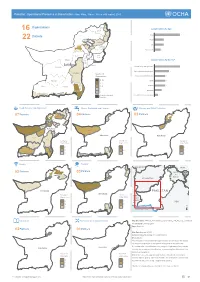
Pakistan: Operational Presence in Balochistan- Who, What, Where, When (4W Matrix) 2019
Pakistan: Operational Presence in Balochistan- Who, What, Where, When (4W matrix) 2019 Organizations Sheerani Organizations by type 16 Zhob Killa NGO 8 Districts Pishin Saifullah Musakhel Killa 22 Abdullah INGO 3 Ziarat Loralai Quetta Harnai UN 3 Barkhan Mastung Sibi Kohlu Government 2 Nushki Kachhi Kalat Lehri Dera Chagai Bugti Kharan Nasirabad Organizations by Sector* Jhal Sohbatpur Magsi Balochistan Jaffarabad Food Security and Agriculture 7 Washuk Water, Sanitation and Hygiene 4 Khuzdar Number of Organizations Women and Child Protection 3 Panjgur 9- 10 Health 3 6- 7 Nutrition 3 Kech Awaran 3- 4 2 2 Las Bela Education 1 Gwadar Drought affected Humanitarian Communications 1 Districts Food Security and Agriculture Water, Sanitation and Hygiene Women and Child Protection Zhob Partners Partners Partners 07 Killa 04 03 Saifullah Pishin Pishin Killa Killa Killa Abdullah Abdullah Abdullah Quetta Quetta Barkhan Sibi Mastung Nushki Nushki Balochistan Chagai Chagai Kharan Jhal Magsi Jaffarabad Balochistan Balochistan Washuk Khuzdar Number of Number of Number of Organizations Organizations Organizations Panjgur 3 3 3 2 2 2 Kech Lasbela Lasbela 1 1 1 Gwadar Health Nutrition Sheerani TURKMENISTAN CHINA Partners Killa Partners Jammu 03 Saifullah 03 Musakhel Aksai Pishin Killa ! ! ! ! ! ! Chin Abdullah ! & ! Loralai ! Quetta Quetta ! Kashmir ! AFGHANISTAN ! ! Chagai Chagai Balochistan PA K I S TA N Balochistan Number of Number of IRAN Organizations Organizations (Islamic Republic 3 3 of) INDIA 2 2 1 1 Lasbela A r a b i a n S e a ± Education Humanitarian