Triphosphate RNA Regulates RIG-I Sensitivity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supporting Information
Supporting Information Celhar et al. 10.1073/pnas.1507052112 SI Materials and Methods using a Nanodrop spectrophotometer (Thermo Fisher Scien- Proteinuria. Proteinuria was assessed using Albustix (Bayer). Al- tific). A TaqMan RNA-to-CT 1-Step Kit (Applied Biosystems) bumin levels in urine were assayed using an Albumin Mouse was used to perform the reverse transcription and quantitative ELISA Kit (Abcam) according to the manufacturer’s instructions; PCR reactions according to the manufacturer’s instructions samples were assayed at a dilution of 1:400. Samples were nor- using TaqMan gene expression assays (Applied Biosystems) to malized for creatinine using a Creatinine (urinary) Colorimetric either Tlr7 (Mm00446590) or the B2m housekeeping gene Assay Kit (Cayman Chemical) according to the manufacturer’s (Mm00437762). Real-time PCR was performed on the 7900H instructions; initial sample dilution of 1:10. fast real-time PCR system and analyzed using SDS 2.4 (Applied Biosystems). Relative mRNA expression was calculated using the Cell Sorting, RNA Isolation, and RT-PCR. Splenic B cells were comparative C method. + − + + t sorted as live CD45 Gr1 B220 CD19 , splenic T cells as live + − + + CD45 Gr1 CD3 CD5 and peritoneal macrophages as live Imaging. Kidney sections from OCT embedded tissue were fixed + − CD45 Gr1 CD11bhiF4/80hi. Sorted cells were centrifuged, re- with 4% paraformaldehyde before permeabilization with acetone suspended in TRIzol (Life Technologies) and stored at −80°. RNA and stained with Phalloidin (AF647) and anti-CD3d (unlabeled was extracted by TRIzol/chloroform and purified with the Qiagen Ab followed by secondary staining with donkey anti-goat Dylight RNeasy Mini purification kit according to the manufacturer’s 550). -

Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis
Virginia Commonwealth University VCU Scholars Compass Theses and Dissertations Graduate School 2010 Molecular Mechanisms Involved Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis Ryan Fassnacht Virginia Commonwealth University Follow this and additional works at: https://scholarscompass.vcu.edu/etd Part of the Physiology Commons © The Author Downloaded from https://scholarscompass.vcu.edu/etd/2246 This Thesis is brought to you for free and open access by the Graduate School at VCU Scholars Compass. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Ryan C. Fassnacht 2010 All Rights Reserved Molecular Mechanisms Involved in the Interaction Effects of HCV and Ethanol on Liver Cirrhosis A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science at Virginia Commonwealth University. by Ryan Christopher Fassnacht, B.S. Hampden Sydney University, 2005 M.S. Virginia Commonwealth University, 2010 Director: Valeria Mas, Ph.D., Associate Professor of Surgery and Pathology Division of Transplant Department of Surgery Virginia Commonwealth University Richmond, Virginia July 9, 2010 Acknowledgement The Author wishes to thank his family and close friends for their support. He would also like to thank the members of the molecular transplant team for their help and advice. This project would not have been possible with out the help of Dr. Valeria Mas and her endearing -

A Functional C-Terminal TRAF3-Binding Site in MAVS Participates in Positive and Negative Regulation of the IFN Antiviral Response
Cell Research (2011) 21:895-910. © 2011 IBCB, SIBS, CAS All rights reserved 1001-0602/11 $ 32.00 npg ORIGINAL ARTICLE www.nature.com/cr A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response Suzanne Paz1, 2, Myriam Vilasco3, Steven J Werden1, Meztli Arguello1, Deshanthe Joseph-Pillai1, 2, Tiejun Zhao1, Thi Lien-Anh Nguyen1, Qiang Sun1, Eliane F Meurs3, Rongtuan Lin1, 4, John Hiscott1, 2, 4 1Terry Fox Molecular Oncology Group, Lady Davis Institute, Jewish General Hospital, Montreal, Quebec H3T1E2, Canada; 2Department of Microbiology and Immunology, McGill University Montreal, Quebec H3A 2B4, Canada; 3Department of Virology, Unit of Hepacivirus and Innate Immunity, Pasteur Institute, Paris 75724 France; 4Department of Medicine, McGill University Montreal, Quebec H3A 2B4, Canada Recognition of viral RNA structures by the cytosolic sensor retinoic acid-inducible gene-I (RIG-I) results in the activation of signaling cascades that culminate with the generation of the type I interferon (IFN) antiviral response. Onset of antiviral and inflammatory responses to viral pathogens necessitates the regulated spatiotemporal recruitment of signaling adapters, kinases and transcriptional proteins to the mitochondrial antiviral signaling protein (MAVS). We previously demonstrated that the serine/threonine kinase IKKε is recruited to the C-terminal region of MAVS following Sendai or vesicular stomatitis virus (VSV) infection, mediated by Lys63-linked polyubiquitination of MAVS at Lys500, resulting in inhibition of downstream IFN signaling (Paz et al, Mol Cell Biol, 2009). In this study, we demonstrate that C-terminus of MAVS harbors a novel TRAF3-binding site in the aa450- 468 region of MAVS. -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Human Ifnα4 / Ifna4 / Interferon Alpha-4 Protein (Fc Tag)
Human IFNα4 / IFNa4 / Interferon alpha-4 Protein (Fc Tag) Catalog Number: 10336-H01H General Information SDS-PAGE: Gene Name Synonym: IFN-alpha4a; INFA4; MGC142200 Protein Construction: A DNA sequence encoding the mature form of human IFN-α4a (NP_066546.1) (Cys 23-Asp 189) was expressed with the N-terminal fused Fc region of human IgG1. Source: Human Expression Host: HEK293 Cells QC Testing Purity: > 97 % as determined by SDS-PAGE Bio Activity: Protein Description Measured in antiviral assays using WISH human amnion cells infected Interferon, alpha 4 (IFNA4) belongs to the alpha/beta interferon family. Two with vesicular stomatitis virus (VSV). The EC50 for this effect is 0.2-1 variants of IFNA4 (IFNA4a and IFNA4b) are known, which differ from each ng/mL. other by changes in their coding regions at nucleotide positions 220 and 410 and can be distinguished by selective restriction enzyme analysis. Endotoxin: Interferons are produced by macrophages, IFN-alpha have antiviral activities. Interferon stimulates the production of two enzymes: a protein < 1.0 EU per μg of the protein as determined by the LAL method kinase and an oligoadenylate synthetase. IFN-alpha, the first cytokine to be produced by recombinant DNA technology, has emerged as an important Stability: regulator of growth and differentiation, affecting cellular communication and Samples are stable for up to twelve months from date of receipt at -70 ℃ signal transduction pathways as well as immunological control. Originally discovered as an antiviral substance, the efficacy of IFN-alpha in malignant, Predicted N terminal: Glu viral, immunological, angiogenic, inflammatory, and fibrotic diseases suggests a spectrum of interrelated pathophysiologies. -

Proquest Dissertations
INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Each original is also photographed in one exposure and is included in reduced form at the back of the book. Photographs included in the original manuscript have been reproduced xerographically in this copy. Higher quality 6” x 9” black and white photographic prints are available for any photographs or illustrations appearing in this copy for an additional charge. Contact UMI directly to order. UMI Bell & Howell Information and Learning 300 North Zeeb Road, Ann Arbor, Ml 48106-1346 USA 800-521-0600 NOTE TO USERS Page(s) missing in number only; text follows. Microfilmed as received. 222-229 This reproduction is the best copy available. UMI MOLECULAR GENETIC AND BIOCHEMICAL STUDIES OF THE HUMAN AND MOUSE MHC COMPLEMENT GENE CLUSTERS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Zhenyu Yang, M.S. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -
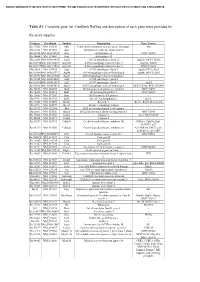
Table S1. Complete Gene List. Genbank Refseq and Description of Each Gene Were Provided By
Document downloaded from http://www.elsevier.es, day 24/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Table S1. Complete gene list. GenBank RefSeq and description of each gene were provided by the array supplier. Unigene GeneBank Symbol Description Gene Name/s Rn.11422 NM_033230 Akt1 V-akt murine thymoma viral oncogene homolog 1 Akt Rn.2104 NM_019288 App Amyloid beta (A4) precursor protein - Rn.23323 NM_001034933 Arsa Arylsulfatase A MGC125207 Rn.94004 NM_033443 Arsb Arylsulfatase B - Rn.6224 NM_001038495 Atg12 ATG12 autophagy related 12 Apg12l, MGC125080 Rn.101734NM_001108809 Atg16l1 ATG16 autophagy related 16-like 1 Apg16l, Wdr30 Rn.104199NM_001191560 Atg16l2 ATG16 autophagy related 16-like 2 RGD1311400 Rn.3084 NM_134394 Atg3 ATG3 autophagy related 3 Apg3l, PIG-1, Pig1 Rn.163086NM_001025711 Atg4b ATG4 autophagy related 4 homolog B Apg4b, MGC112887 Rn.23378 NM_001107948 Atg4c ATG4 autophagy related 4 homolog C - Rn.98385 NM_001014250 Atg5 ATG5 autophagy related 5 - Rn.162765NM_001012097 Atg7 ATG7 autophagy related 7 Apg7l Rn.35248 NM_001014218 Atg9a ATG9 autophagy related 9 homolog A MGC105908, RGD1310450 Rn.36696 NM_022698 Bad BCL2-associated agonist of cell death MGC72439 Rn.14598 NM_053812 Bak1 BCL2-antagonist/killer 1 MGC108627 Rn.10668 NM_017059 Bax Bcl2-associated X protein - Rn.9996 NM_016993 Bcl2 B-cell CLL/lymphoma 2 Bcl-2 Rn.10323 NM_031535 Bcl2l1 Bcl2-like 1 Bcl-xl, Bcl2l, Bclx, bcl-X Rn.2776 NM_053739 Becn1 Beclin 1, autophagy related - Rn.31142 NM_022684 -

Datasheet Blank Template
SAN TA C RUZ BI OTEC HNOL OG Y, INC . DOM3Z (B-12): sc-393141 BACKGROUND APPLICATIONS DOM3Z (dom-3 homolog Z), also known as NG6 or DOM3L, is a 396 amino DOM3Z (B-12) is recommended for detection of DOM3Z of mouse, rat and acid ubiquitously expressed protein belonging to the DOM3Z family. The gene human origin by Western Blotting (starting dilution 1:100, dilution range encoding DOM3Z maps to human chromosome 6 in the major histocompati - 1:100-1:1000), immunoprecipitation [1-2 µg per 100-500 µg of total protein bility complex (MHC) class III region. Chromosome 6 contains 170 million base (1 ml of cell lysate)], immunofluorescence (starting dilution 1:50, dilution pairs and comprises nearly 6% of the human genome. Deletion of a portion range 1:50-1:500), immunohistochemistry (including paraffin-embedded of the q arm of chromosome 6 is associated with early onset intestinal can - sections) (starting dilution 1:50, dilution range 1:50-1:500) and solid phase cer, suggesting the presence of a cancer susceptibility locus. Additionally, ELISA (starting dilution 1:30, dilution range 1:30-1:3000). Porphyria cutanea tarda, Parkinson’s disease and Stickler syndrome are all Suitable for use as control antibody for DOM3Z siRNA (h): sc-95321, DOM3Z associated with genes that map to chromosome 6. siRNA (m): sc-143143, DOM3Z shRNA Plasmid (h): sc-95321-SH, DOM3Z shRNA Plasmid (m): sc-143143-SH, DOM3Z shRNA (h) Lentiviral Particles: REFERENCES sc-95321-V and DOM3Z shRNA (m) Lentiviral Particles: sc-143143-V. 1. Brunner, H.G., et al . -

DNA Methylation Alterations in Blood Cells of Toddlers with Down Syndrome
G C A T T A C G G C A T genes Article DNA Methylation Alterations in Blood Cells of Toddlers with Down Syndrome Oxana Yu. Naumova 1,2,* , Rebecca Lipschutz 2, Sergey Yu. Rychkov 1, Olga V. Zhukova 1 and Elena L. Grigorenko 2,3,4,* 1 Vavilov Institute of General Genetics RAS, 119991 Moscow, Russia; [email protected] (S.Y.R.); [email protected] (O.V.Z.) 2 Department of Psychology, University of Houston, Houston, TX 77204, USA; [email protected] 3 Department of Psychology, Saint-Petersburg State University, 199034 Saint Petersburg, Russia 4 Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA * Correspondence: [email protected] or [email protected] (O.Y.N.); [email protected] (E.L.G.) Abstract: Recent research has provided evidence on genome-wide alterations in DNA methylation patterns due to trisomy 21, which have been detected in various tissues of individuals with Down syndrome (DS) across different developmental stages. Here, we report new data on the systematic genome-wide DNA methylation perturbations in blood cells of individuals with DS from a previously understudied age group—young children. We show that the study findings are highly consistent with those from the prior literature. In addition, utilizing relevant published data from two other developmental stages, neonatal and adult, we track a quasi-longitudinal trend in the DS-associated DNA methylation patterns as a systematic epigenomic destabilization with age. Citation: Naumova, O.Y.; Lipschutz, R.; Rychkov, S.Y.; Keywords: Down syndrome; infants and toddlers; trisomy 21; DNA methylation; Illumina 450K Zhukova, O.V.; Grigorenko, E.L. -

Interferonsource.Com What's Inside New Products Research Tools Protocols & Tips Product Citations What's Your IFN-Α
Interferon Matters A quarterly newsletter by PBL InterferonSource interferonsource.com May 2007, Issue 2 u What’s Inside What’s Your IFN-a Type? New Products Are all Type I IFNs created equal? Of Mice and Men page 4 & 5 Type I Interferons were the first cytokines to be The biological significance of the expression of discovered. They are a family of homologous cytokines so many different IFN-a subtypes has yet to be Rhesus/Cynomolgus originally identified for their antiviral activity but have determined. However, reports suggest that they show IFN-a2 Subtype also been shown to have both anti-proliferative and quantitatively distinct anti-viral, anti-proliferative, immunomodulatory activities. In humans and mice and killer cell-stimulatory activities. It is therefore Mouse IFN-a4 Subtype they are encoded by an intronless multigene family of importance for researchers to characterize and clustered on human chromosome 9 and murine study these IFN-a subtypes and their possible post- chromosome 4 respectively. translational modifications. Research Tools Also in both species there are multiple Interferon- The common models for IFN-a subtypes study are page 6 & 7 Alpha (IFN-a) genes and one Interferon-Beta (IFN-b) human and mouse. In 1998, Lawson et al showed that Human IFN-a gene. Other Type I Interferon genes described include there are in vivo differences in the antiviral activity of the Interferon-Omega (IFN-w) gene, murine limitin the mouse IFN-a subtypes.1 Using an experimental Multi-Subtype ELISA Kit gene, as well as the human/murine Interferon-Epilson, mouse model of IFN transgene expression in vivo, TM Interferon-Kappa, and Interferon-Tau (IFN-e, IFN-k, and the authors showed that mouse IFN-a1 has a greater iLite Human IFN-a Kit IFN-t).