Asparagine Biosynthesis and Utilization
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

Proline- and Alanine-Rich N-Terminal Extension of the Basic Bovine P-Crystallin B1 Chains
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Volume 161, number 2 FEBS 0790 September 1983 Proline- and alanine-rich N-terminal extension of the basic bovine P-crystallin B1 chains G.A.M. Berbers, W.A. Hoekman, H. Bloemendal, W.W. de Jong, T. Kleinschmidt* and G. Braunitzer* Laboratorium voor Biochemie, Universiteit van Nijmegen, Geert Grooteplein Noord 21, 6525 EZ Nijmegen, The Netherlands and *Max-Planck-Institut ft’ir Biochemie, Abteilung Proteinchemie, D-8033 Martinsried bei Miinchen, FRG Received 22 June 1983 The amino acid sequence of the N-terminal region of the two basic bovine &crystallin B1 chains has been analyzed. The results reveal that @Bibis derived in vivo from the primary gene product @la by removal of a short N-terminal sequence. It appears that them1 chains have the same domain structure as observed in other /3- and y-crystallin chains. They have, however, a very long N-terminal extension in comparison with other &chains. This extension is mainly composed of a remarkable Pro- and Ala-rich sequence, which suggests an interaction of these structural proteins with the cytoskeleton and/or the plasma membranes of the lens cells. Protein sequence Bovine &crystallin N-terminal extension Proline- and aianine-rich Domain structure 1. INTRODUCTION 33 000 and 31000, respectively, are characteristic for fltikt, [ 111. ,8Fha is a primary gene product, from The crystallins are evolutionary highly conserv- which ,8&b arises by post-translational modifica- ed structural eye lens proteins, which can be divid; tion [lo-121, most probably a proteolytic step ed into 4 classes: (Y-,,&, y- and t-crystallin [ 11. -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

An Integrated Meta-Analysis of Peripheral Blood Metabolites and Biological Functions in Major Depressive Disorder
Molecular Psychiatry https://doi.org/10.1038/s41380-020-0645-4 ARTICLE An integrated meta-analysis of peripheral blood metabolites and biological functions in major depressive disorder 1,2,3 1,2,3 1,2,3 1,3 1,3 4,5 1,3 1,3 Juncai Pu ● Yiyun Liu ● Hanping Zhang ● Lu Tian ● Siwen Gui ● Yue Yu ● Xiang Chen ● Yue Chen ● 1,2,3 1,3 1,3 1,3 1,3 1,2,3 Lining Yang ● Yanqin Ran ● Xiaogang Zhong ● Shaohua Xu ● Xuemian Song ● Lanxiang Liu ● 1,2,3 1,3 1,2,3 Peng Zheng ● Haiyang Wang ● Peng Xie Received: 3 June 2019 / Revised: 24 December 2019 / Accepted: 10 January 2020 © The Author(s) 2020. This article is published with open access Abstract Major depressive disorder (MDD) is a serious mental illness, characterized by high morbidity, which has increased in recent decades. However, the molecular mechanisms underlying MDD remain unclear. Previous studies have identified altered metabolic profiles in peripheral tissues associated with MDD. Using curated metabolic characterization data from a large sample of MDD patients, we meta-analyzed the results of metabolites in peripheral blood. Pathway and network analyses were then performed to elucidate the biological themes within these altered metabolites. We identified 23 differentially 1234567890();,: 1234567890();,: expressed metabolites between MDD patients and controls from 46 studies. MDD patients were characterized by higher levels of asymmetric dimethylarginine, tyramine, 2-hydroxybutyric acid, phosphatidylcholine (32:1), and taurochenode- soxycholic acid and lower levels of L-acetylcarnitine, creatinine, L-asparagine, L-glutamine, linoleic acid, pyruvic acid, palmitoleic acid, L-serine, oleic acid, myo-inositol, dodecanoic acid, L-methionine, hypoxanthine, palmitic acid, L-tryptophan, kynurenic acid, taurine, and 25-hydroxyvitamin D compared with controls. -

Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat
Pediat. Res. 6: 713-719 (1972) Amino acid neonate intestine transport, amino acid Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat J. F. FITZGERALD1431, S. REISER, AND P. A. CHRISTIANSEN Departments of Pediatrics, Medicine, and Biochemistry, and Gastrointestinal Research Laboratory, Indiana University School of Medicine and Veterans Administration Hospital, Indianapolis, Indiana, USA Extract The activity of amino acid transport pathways in the small intestine of the 2-day-old rat was investigated. Transport was determined by measuring the uptake of 1 mM con- centrations of various amino acids by intestinal segments after a 5- or 10-min incuba- tion and it was expressed as intracellular accumulation. The neutral amino acid transport pathway was well developed with intracellular accumulation values for leucine, isoleucine, valine, methionine, tryptophan, phenyl- alanine, tyrosine, and alanine ranging from 3.9-5.6 mM/5 min. The intracellular accumulation of the hydroxy-containing neutral amino acids threonine (essential) and serine (nonessential) were 2.7 mM/5 min, a value significantly lower than those of the other neutral amino acids. The accumulation of histidine was also well below the level for the other neutral amino acids (1.9 mM/5 min). The basic amino acid transport pathway was also operational with accumulation values for lysine, arginine and ornithine ranging from 1.7-2.0 mM/5 min. Accumulation of the essential amino acid lysine was not statistically different from that of nonessential ornithine. Ac- cumulation of aspartic and glutamic acid was only 0.24-0.28 mM/5 min indicating a very low activity of the acidic amino acid transport pathway. -
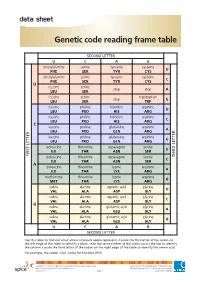
Genetic Code Reading Frame Table
data sheet Genetic code reading frame table SECOND LETTER U C A G phenylalanine serine tyrosine cysteine U PHE SER TYR CYS phenylalanine serine tyrosine cysteine C PHE SER TYR CYS U leucine serine stop stop A LEU SER leucine serine tryptophan stop G LEU SER TRP leucine proline histidine arginine U LEU PRO HIS ARG leucine proline histidine arginine C LEU PRO HIS ARG C leucine proline glutamine arginine A LEU PRO GLN ARG leucine proline glutamine arginine G LEU PRO GLN ARG isoleucine threonine asparagine serine U ILE THR ASN SER FIRST LETTER isoleucine threonine asparagine serine LETTERTHIRD C ILE THR ASN SER A isoleucine threonine lysine arginine A ILE THR LYS ARG methionine threonine lysine arginine G MET THR LYS ARG valine alanine aspartic acid glycine U VAL ALA ASP GLY valine alanine aspartic acid glycine C VAL ALA ASP GLY G valine alanine glutamic acid glycine A VAL ALA GLU GLY valine alanine glutamic acid glycine G VAL ALA GLU GLY U C A G SECOND LETTER Use this table to find out what amino acid each codon represents. Locate the first letter of the codon on the left edge of the table to identify a block, then the second letter of the codon across the top to identify the column. Locate the third letter of the codon on the right edge of the table to identify the amino acid. For example, the codon ‘CAU’ codes for histidine (HIS). ast0812 | Proteins 3: Genetic code reading frame table (data sheet) developed for the Department of Education WA © The University of Western Australia 2012 for conditions of use see spice.wa.edu.au/usage version 1.0 page 1 Licensed for NEALS. -

Analysis of Amino Acids by HPLC
Analysis of Amino Acids by HPLC Rita Steed Agilent Technologies, Inc. 800-227-9770 opt 3/opt3/opt 2 Amino Acid Analysis - Agilent Restricted Page 1 June 24, 2010 Outline • Amino Acids – Structure, Chemistry • Separation Considerations • Challenges • Instrumentation • Derivatization – OPA, FMOC • Overview of Separations • Examples Amino Acid Analysis - Agilent Restricted Page 2 June 24, 2010 Amino Acids – Structure, Chemistry CH3 Alanine (()Ala) Glutamic Acid (()Glu) Amino Acid Analysis - Agilent Restricted Page 3 June 24, 2010 Amino Acids – Zwitterionic Amino Acid Analysis - Agilent Restricted Page 4 June 24, 2010 Separation Considerations • Zwitterions - poor solubility near iso- …electric point • Most have poor UV absorbance • Derivatization – OPA, FMOC •Reduce polarity – increases retention in reversed-phase chromatoggpyraphy •Improve sensitivity – UV, Fluorescence • Detector; DAD, FLD, MS, ELSD Amino Acid Analysis - Agilent Restricted Page 5 June 24, 2010 Ortho Phthalaldehyde (OPA) and Fluorenylmethoxy chloroformate (FMOC) Reactions with Amines OPA O SR’ R’SH H NR +RNH2 H Room Temperature O Fluorescence: Ex 340nm, Em 450nm Non-fluorescent DAD: 338 , 10nm; Ref . 390 , 20nm Does not absorb at 338nm FMOC RR’NH - HCl + or Room Temperature RNH2 NRR’ or NHR Fluorescence: Ex 260nm, Em 325nm Fluorescent DAD: 262, 16nm; Ref. 324,8nm Absorbs at 262nm and Fluorescences at 324nm Group/Presentation Title Agilent Restricted Month ##, 200X Names and Order of Elution for OPA and FMOC Derivatives of Amino Acids Peak # AA Name AA Abbreviation Derivative Type Peak # AA Name AA Abbreviation Derivative Type Group/Presentation Title Agilent Restricted Month ##, 200X Agilent AAA Methods - They’ve Evolved • Automated Amino Acid Analysis – AminoQuant I & II (1987) •1090 •1100, Pub. -

Amino Acid Degradation
BI/CH 422/622 OUTLINE: OUTLINE: Protein Degradation (Catabolism) Digestion Amino-Acid Degradation Inside of cells Protein turnover Dealing with the carbon Ubiquitin Fates of the 29 Activation-E1 Seven Families Conjugation-E2 nitrogen atoms in 20 1. ADENQ Ligation-E3 AA: Proteosome 2. RPH 9 ammonia oxidase Amino-Acid Degradation 18 transamination Ammonia 2 urea one-carbon metabolism free transamination-mechanism to know THF Urea Cycle – dealing with the nitrogen SAM 5 Steps Carbamoyl-phosphate synthetase 3. GSC Ornithine transcarbamylase PLP uses Arginino-succinate synthetase Arginino-succinase 4. MT – one carbon metabolism Arginase 5. FY – oxidase vs oxygenase Energetics Urea Bi-cycle 6. KW – Urea Cycle – dealing with the nitrogen 7. BCAA – VIL Feeding the Urea Cycle Glucose-Alanine Cycle Convergence with Fatty acid-odd chain Free Ammonia Overview Glutamine Glutamate dehydrogenase Overall energetics Amino Acid A. Concepts 1. ConvergentDegradation 2. ketogenic/glucogenic 3. Reactions seen before The SEVEN (7) Families B. Transaminase (A,D,E) / Deaminase (Q,N) Family C. Related to biosynthesis (R,P,H; C,G,S; M,T) 1.Glu Family a. Introduce oxidases/oxygenases b. Introduce one-carbon metabolism (1C) 2.Pyruvate Family a. PLP reactions 3. a-Ketobutyric Family (M,T) a. 1-C metabolism D. Dedicated 1. Aromatic Family (F,Y) a. oxidases/oxygenases 2. a-Ketoadipic Family (K,W) 3. Branched-chain Family (V,I,L) E. Convergence with Fatty Acids: propionyl-CoA 29 N 1 Amino Acid Degradation • Intermediates of the central metabolic pathway • Some amino acids result in more than one intermediate. • Ketogenic amino acids can be converted to ketone bodies. -

Metabolic Classification of the Amino Acids
Metabolic Classification of the Amino Acids *Essential and Non -essential * Glucogenic and Ketogenic 1 Essential Amino Acids • Of the 20 amino acids that make up proteins 10 of them can be synthesized by the human body • The other 10 amino acids must be acquired from food sources. These amino acids are known as essential amino acids 2 Essential Amino Acids Complete protein Incomplete protein • Contains all 10 Lack one of more of the essential amino acids essential amino acids • Proteins derived from Most vegetable proteins animal sources are are incomplete proteins complete proteins Beans are an exception to • Beans contain some this generalizations complete protein as well 3 Essential Amino Acids in Humans • Required in diet • Humans incapable of forming requisite carbon skeleton • Arginine* • Lysine • Histidine* • Methionine • Isoleucine • Threonine • Leucine • Phenylalanine • Valine • Tryptophan * Essential in children, not in adults 4 Non-Essential Amino Acids in Humans • Not required in diet • Can be formed from ααα-keto acids by transamination and subsequent reactions • Alanine • Glycine • Asparagine • Proline • Aspartate • Serine • Glutamate • Cysteine (from Met *) • Glutamine • Tyrosine (from Phe *) * Essential amino acids 5 Essential and Nonessential Amino Acids Nonessential Essentia l Alanine Arginine * Asparagine Histidine * Aspartate Isoleucine Cysteine Leucine Glutamate Lysine Glutamine Methionine Glycine Phenylalanine Proline Threonine Serine Tyrptophan Tyrosine Valine Amino acids are classified as glucogenic or ketogenic -

Safety Assessment of Α-Amino Acids As Used in Cosmetics
Safety Assessment of α-Amino Acids as Used in Cosmetics Status: Final Report for Public Distribution Release Date: October 5, 2012 Panel Meeting Date: September 10-11, 2012 The 2012 Cosmetic Ingredient Review Expert Panel members are: Chairman, Wilma F. Bergfeld, M.D., F.A.C.P.; Donald V. Belsito, M.D.; Curtis D. Klaassen, Ph.D.; Daniel C. Liebler, Ph.D.; Ronald A. Hill, Ph.D. James G. Marks, Jr., M.D.; Ronald C. Shank, Ph.D.; Thomas J. Slaga, Ph.D.; and Paul W. Snyder, D.V.M., Ph.D. The CIR Director is F. Alan Andersen, Ph.D. This safety assessment was prepared by Christina L. Burnett, Scientific Analyst/Writer, and Bart Heldreth, Ph.D., Chemist CIR. © Cosmetic Ingredient Review 1101 17th Street, NW, Suite 412 Washington, DC 20036-4702 ph 202.331.0651 fax 202.331.0088 [email protected] 1 ABSTRACT The Cosmetic Ingredient Review Expert Panel (the Panel) reviewed the safety of α-amino acids, which function primarily as hair and skin conditioning agents in cosmetic products. The safety of α-amino acids as direct food additives has been well established based on extensive research through acute and chronic dietary exposures. The Panel focused its review on dermal irritation and sensitization data relevant to the use of these ingredients in topical cosmetics. The Panel concluded that α-amino acids were safe as cosmetic ingredients in the practices of use and concentration of this safety assessment. INTRODUCTION Amino acids and their salts are widely used as cosmetic ingredients, and function primarily as hair conditioning agents and skin conditioning agents (humectant and miscellaneous). -

Amino Acid Catabolism
Amino Acid Catabolism • Dietary Proteins • Turnover of Protein • Cellular protein • Deamination • Urea cycle • Carbon skeletons of amino acids Amino Acid Metabolism •Metabolism of the 20 common amino acids is considered from the origins and fates of their: (1) Nitrogen atoms (2) Carbon skeletons •For mammals: Essential amino acids must be obtained from diet Nonessential amino acids - can be synthesized Amino Acid Catabolism • Amino acids from degraded proteins or from diet can be used for the biosynthesis of new proteins • During starvation proteins are degraded to amino acids to support glucose formation • First step is often removal of the α-amino group • Carbon chains are altered for entry into central pathways of carbon metabolism Dietary Proteins • Digested in intestine • by peptidases • transport of amino acids • active transport coupled with Na+ Protein Turnover • Proteins are continuously synthesized and degraded (turnover) (half-lives minutes to weeks) • Lysosomal hydrolysis degrades some proteins • Some proteins are targeted for degradation by a covalent attachment (through lysine residues) of ubiquitin (C terminus) • Proteasome hydrolyzes ubiquitinated proteins Turnover of Protein • Cellular protein • Proteasome degrades protein with Ub tags • T 1/2 determined by amino terminus residue • stable: ala, pro, gly, met greater than 20h • unstable: arg, lys, his, phe 2-30 min Ubibiquitin • Ubiquitin protein, 8.5 kD • highly conserved in yeast/humans • carboxy terminal attaches to ε-lysine amino group • Chains of 4 or more Ub molecules -

Plasma Concentrations and Intakes of Amino Acids in Male Meat-Eaters, fish-Eaters, Vegetarians and Vegans: a Cross-Sectional Analysis in the EPIC-Oxford Cohort
European Journal of Clinical Nutrition (2016) 70, 306–312 OPEN © 2016 Macmillan Publishers Limited All rights reserved 0954-3007/16 www.nature.com/ejcn ORIGINAL ARTICLE Plasma concentrations and intakes of amino acids in male meat-eaters, fish-eaters, vegetarians and vegans: a cross-sectional analysis in the EPIC-Oxford cohort JA Schmidt1, S Rinaldi2, A Scalbert2, P Ferrari2, D Achaintre2, MJ Gunter3, PN Appleby1, TJ Key1 and RC Travis1 BACKGROUND/OBJECTIVES: We aimed to investigate the differences in plasma concentrations and in intakes of amino acids between male meat-eaters, fish-eaters, vegetarians and vegans in the Oxford arm of the European Prospective Investigation into Cancer and Nutrition. SUBJECTS/METHODS: This cross-sectional analysis included 392 men, aged 30–49 years. Plasma amino acid concentrations were measured with a targeted metabolomic approach using mass spectrometry, and dietary intake was assessed using a food frequency questionnaire. Differences between diet groups in mean plasma concentrations and intakes of amino acids were examined using analysis of variance, controlling for potential confounding factors and multiple testing. RESULTS: In plasma, concentrations of 6 out of 21 amino acids varied significantly by diet group, with differences of − 13% to +16% between meat-eaters and vegans. Concentrations of methionine, tryptophan and tyrosine were highest in fish-eaters and vegetarians, followed by meat-eaters, and lowest in vegans. A broadly similar pattern was seen for lysine, whereas alanine concentration was highest in fish-eaters and lowest in meat-eaters. For glycine, vegans had the highest concentration and meat-eaters the lowest. Intakes of all 18 dietary amino acids differed by diet group; for the majority of these, intake was highest in meat-eaters followed by fish-eaters, then vegetarians and lowest in vegans (up to 47% lower than in meat-eaters).