Dlx5 Regulation of Craniofacial Development 3833
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

INDEX Abducent Neurons Anatomy 135 Clinical Signs 137 Diseases
INDEX Abducent neurons Anatomy 135 Clinical signs 137 Diseases 139 Function 135 Abiotrophic sensorineural deafness 438 Abiotrophy 100, 363 Auditory 438 Cerebellar cortical 363 Motor neuron 100 Nucleus ambiguus 159 Peripheral vestibular 336 Abscess-Brainstem 330 Caudal cranial fossa 343 Cerebellar 344 Cerebral 416, 418 Pituitary 162 Streptococcus equi 418 Abyssian cat-Glucocerebrosidosis 427 Myastheina gravis 93 Accessory neurons Anatomy 152 Clinical signs 153 Diseases 153 Acetozolamide 212 Acetylcholine 78, 169, 354, 468, 469 Receptor 78 Acetylcholinesterase 79 Achiasmatic Belgian sheepdog 345 Acoustic stria 434 Acral mutilation 237 Adenohypophysis 483 Releasing factors 483 Adenosylmethionine 262 Adhesion-Interthalamic 33, 476 Adiposogenital syndrome 484 Adipsia 458, 484 Adrenocorticotrophic hormone 485 Adversive syndrome 72, 205, 460 Afghan hound-Inherited myelinolytic encephalomyelopathy 264 Agenized flour 452 Aino virus 44 Akabane virus 43 Akita-Congenital peripiheral vestibular disease 336 Alaskan husky encephalopathy 522 Albinism, ocular 345 Albinotic sensorineural deafness 438 Alexander disease 335 Allodynia 190 Alpha fucosidosis 427 Alpha glucosidosis 427 Alpha iduronidase 427 Alpha mannosidase 427 Alpha melanotropism 484 Alsatian-idiopathic epilepsy 458 Alternative anticonvulsant drugs 466 American Bulldog-Ceroid lipofuscinosis 385, 428 American Miniature Horse-Narcolepsy 470 American StaffordshireTerrier-Cerebellar cortical abiotrophy 367 Amikacin 329 Aminocaproic acid 262 Aminoglycoside antibiotics 329, 439 Amprolium toxicity -

Multifactorial Erβ and NOTCH1 Control of Squamous Differentiation and Cancer
Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer Yang Sui Brooks, … , Karine Lefort, G. Paolo Dotto J Clin Invest. 2014;124(5):2260-2276. https://doi.org/10.1172/JCI72718. Research Article Oncology Downmodulation or loss-of-function mutations of the gene encoding NOTCH1 are associated with dysfunctional squamous cell differentiation and development of squamous cell carcinoma (SCC) in skin and internal organs. While NOTCH1 receptor activation has been well characterized, little is known about how NOTCH1 gene transcription is regulated. Using bioinformatics and functional screening approaches, we identified several regulators of the NOTCH1 gene in keratinocytes, with the transcription factors DLX5 and EGR3 and estrogen receptor β (ERβ) directly controlling its expression in differentiation. DLX5 and ERG3 are required for RNA polymerase II (PolII) recruitment to the NOTCH1 locus, while ERβ controls NOTCH1 transcription through RNA PolII pause release. Expression of several identified NOTCH1 regulators, including ERβ, is frequently compromised in skin, head and neck, and lung SCCs and SCC-derived cell lines. Furthermore, a keratinocyte ERβ–dependent program of gene expression is subverted in SCCs from various body sites, and there are consistent differences in mutation and gene-expression signatures of head and neck and lung SCCs in female versus male patients. Experimentally increased ERβ expression or treatment with ERβ agonists inhibited proliferation of SCC cells and promoted NOTCH1 expression and squamous differentiation both in vitro and in mouse xenotransplants. Our data identify a link between transcriptional control of NOTCH1 expression and the estrogen response in keratinocytes, with implications for differentiation therapy of squamous cancer. Find the latest version: https://jci.me/72718/pdf Research article Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer Yang Sui Brooks,1,2 Paola Ostano,3 Seung-Hee Jo,1,2 Jun Dai,1,2 Spiro Getsios,4 Piotr Dziunycz,5 Günther F.L. -

Comparative Analysis of a Teleost Skeleton Transcriptome Provides Insight Into Its Regulation
Accepted Manuscript Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation Florbela A. Vieira, M.A.S. Thorne, K. Stueber, M. Darias, R. Reinhardt, M.S. Clark, E. Gisbert, D.M. Power PII: S0016-6480(13)00264-5 DOI: http://dx.doi.org/10.1016/j.ygcen.2013.05.025 Reference: YGCEN 11541 To appear in: General and Comparative Endocrinology Please cite this article as: Vieira, F.A., Thorne, M.A.S., Stueber, K., Darias, M., Reinhardt, R., Clark, M.S., Gisbert, E., Power, D.M., Comparative analysis of a teleost skeleton transcriptome provides insight into its regulation, General and Comparative Endocrinology (2013), doi: http://dx.doi.org/10.1016/j.ygcen.2013.05.025 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 Comparative analysis of a teleost skeleton transcriptome 2 provides insight into its regulation 3 4 Florbela A. Vieira1§, M. A. S. Thorne2, K. Stueber3, M. Darias4,5, R. Reinhardt3, M. 5 S. Clark2, E. Gisbert4 and D. M. Power1 6 7 1Center of Marine Sciences, Universidade do Algarve, Faro, Portugal. 8 2British Antarctic Survey – Natural Environment Research Council, High Cross, 9 Madingley Road, Cambridge, CB3 0ET, UK. -

¬LACZ-REPORTER MAPPING of Dlx5/6 EXPRESSION AND
Received: 26 December 2019 Revised: 10 May 2020 Accepted: 11 May 2020 DOI: 10.1002/cne.24952 RESEARCH ARTICLE LacZ-reporter mapping of Dlx5/6 expression and genoarchitectural analysis of the postnatal mouse prethalamus Luis Puelles1 | Carmen Diaz2 | Thorsten Stühmer3 | José L. Ferran1 | Margaret Martínez-de la Torre1 | John L. R. Rubenstein3 1Department of Human Anatomy and Psychobiology and IMIB-Arrixaca Institute, Abstract University of Murcia, Murcia, Spain We present here a thorough and complete analysis of mouse P0-P140 prethalamic 2 Department of Medical Sciences, School of histogenetic subdivisions and corresponding nuclear derivatives, in the context of Medicine and Institute for Research in Neurological Disabilities, University of Castilla- local tract landmarks. The study used as fundamental material brains from a transgenic La Mancha, Albacete, Spain mouse line that expresses LacZ under the control of an intragenic enhancer of Dlx5 3Nina Ireland Laboratory of Developmental Neurobiology, Department of Psychiatry, and Dlx6 (Dlx5/6-LacZ). Subtle shadings of LacZ signal, jointly with pan-DLX immu- UCSF Medical School, San Francisco, noreaction, and several other ancillary protein or RNA markers, including Calb2 and California Nkx2.2 ISH (for the prethalamic eminence, and derivatives of the rostral zona limitans Correspondence shell domain, respectively) were mapped across the prethalamus. The resulting model Luis Puelles, Department of Human Anatomy and Psychobiology, School of Medicine, of the prethalamic region postulates tetrapartite rostrocaudal and dorsoventral subdi- University of Murcia, Murcia 30071, Spain. visions, as well as a tripartite radial stratification, each cell population showing a char- Email: [email protected] acteristic molecular profile. Some novel nuclei are proposed, and some instances of Carmen Díaz, Department of Medical potential tangential cell migration were noted. -

Molecular Mechanisms Regulating Mammalian Forebrain Development
Molecular Mechanisms Regulating Mammalian Forebrain Development By David Chun Cheong Tsui A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Institute of Medical Science University of Toronto © Copyright by David Chun Cheong Tsui (2013) Title: Molecular Mechanisms Regulating Mammalian Forebrain Development Name: David Chun Cheong Tsui Degree: Doctor of Philosophy, 2013 Department: Institute of Medical Sciences, University of Toronto ABSTRACT While the extrinsic factors regulating neurogenesis in the developing forebrain have been widely studied, the mechanisms downstream of the various signaling pathways are relatively ill-defined. In particular, we focused on proteins that have been implicated in cognitive dysfunction. Here, we ask what role two cell intrinsic factors play in the development of two different neurogenic compartments in the forebrain. In the first part of the thesis, the transcription factor FoxP2, which is mutated in individuals who have specific language deficits, was identified to regulate neurogenesis in the developing cortex, in part by regulating the transition from the radial precursors to the transit amplifying intermediate progenitors. Moreover, we found that ectopic expression of the human homologue of the protein promotes neurogenesis in the murine cortex, thereby acting as a gain-of-function isoform. In the second part of the thesis, the histone acetyltransferase CREB-binding protein (CBP) was identified as regulating the generation of neurons from medial ganglionic eminence precursors, similar to its role in the developing cortex. But CBP also plays a more substantial role in the expression of late interneuron markers, suggesting that it is continuously required for the various stages of neurogenesis at least in the ventral neurogenic niche. -
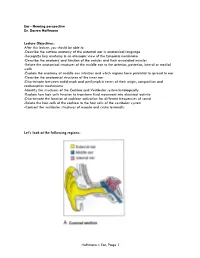
Ear, Page 1 Lecture Outline
Ear - Hearing perspective Dr. Darren Hoffmann Lecture Objectives: After this lecture, you should be able to: -Describe the surface anatomy of the external ear in anatomical language -Recognize key anatomy in an otoscopic view of the tympanic membrane -Describe the anatomy and function of the ossicles and their associated muscles -Relate the anatomical structures of the middle ear to the anterior, posterior, lateral or medial walls -Explain the anatomy of middle ear infection and which regions have potential to spread to ear -Describe the anatomical structures of the inner ear -Discriminate between endolymph and perilymph in terms of their origin, composition and reabsorption mechanisms -Identify the structures of the Cochlea and Vestibular system histologically -Explain how hair cells function to transform fluid movement into electrical activity -Discriminate the location of cochlear activation for different frequencies of sound -Relate the hair cells of the cochlea to the hair cells of the vestibular system -Contrast the vestibular structures of macula and crista terminalis Let’s look at the following regions: Hoffmann – Ear, Page 1 Lecture Outline: C1. External Ear Function: Amplification of Sound waves Parts Auricle Visible part of external ear (pinna) Helix – large outer rim Tragus – tab anterior to external auditory meatus External auditory meatus Auditory Canal/External Auditory Meatus Leads from Auricle to Tympanic membrane Starts cartilaginous, becomes bony as it enters petrous part of temporal bone Earwax (Cerumen) Complex mixture -

Exclusion of Dlx5/6 Expression from the Distal-Most Mandibular Arches Enables BMP-Mediated Specification of the Distal Cap
Exclusion of Dlx5/6 expression from the distal-most mandibular arches enables BMP-mediated specification of the distal cap Joshua W. Vincentza, Jose J. Casasnovasa, Ralston M. Barnesb, Jianwen Quec, David E. Clouthierd, Jun Wange, and Anthony B. Firullia,1 aRiley Heart Research Center, Herman B. Wells Center for Pediatric Research Division of Pediatric Cardiology, Departments of Anatomy, Biochemistry, and Medical and Molecular Genetics, Indiana University Medical School, Indianapolis, IN 46202; bBiologics Discovery California, Bristol-Myers Squibb, Redwood City, CA 94063; cDepartment of Medicine, Columbia University Medical Center, New York, NY 10032; dDepartment of Craniofacial Biology, University of Colorado Anschutz Medical Campus, Aurora, CO 80108; and eDepartment of Molecular Physiology and Biophysics, Texas Heart Institute, Baylor College of Medicine, Houston, TX 77030 Edited by Clifford J. Tabin, Harvard Medical School, Boston, MA, and approved May 26, 2016 (received for review March 8, 2016) Cranial neural crest cells (crNCCs) migrate from the neural tube to Smads, H2, and GATA2/3 provide positive transcriptional inputs the pharyngeal arches (PAs) of the developing embryo and, sub- that serve to counteract the activity of Dlx proteins, thereby re- sequently, differentiate into bone and connective tissue to form the lieving EDN1-meditated repression. Together, these findings in- mandible. Within the PAs, crNCCs respond to local signaling cues to tegrate the communication between BMP and EDN1 signaling partition into the proximo-distally oriented subdomains that convey that establishes the distal cap of the mandible. positional information to these developing tissues. Here, we show that the distal-most of these subdomains, the distal cap, is marked Results by expression of the transcription factor Hand1 (H1) and gives rise to The H1Cre Lineage Gives Rise to the Lower Incisors, in a HAND Factor- the ectomesenchymal derivatives of the lower incisors. -

The Roles of RNA Polymerase I and III Subunits Polr1a, Polr1c, and Polr1d in Craniofacial Development BY
The roles of RNA Polymerase I and III subunits Polr1a, Polr1c, and Polr1d in craniofacial development BY © 2016 Kristin Emily Noack Watt Submitted to the graduate degree program in Anatomy and Cell Biology and to the Graduate Faculty of The University of Kansas Medical Center in partial fulfillment of the requirements for the degree of Doctor of Philosophy. Paul Trainor, Co-Chairperson Brenda Rongish, Co-Chairperson Brian Andrews Jennifer Gerton Tatjana Piotrowski Russell Swerdlow Date Defended: January 26, 2016 The Dissertation Committee for Kristin Watt certifies that this is the approved version of the following dissertation: The roles of RNA Polymerase I and III subunits Polr1a, Polr1c, and Polr1d in craniofacial development Paul Trainor, Co-Chairperson Brenda Rongish, Co-Chairperson Date approved: February 2, 2016 ii Abstract Craniofacial anomalies account for approximately one-third of all birth defects. Two examples of syndromes associated with craniofacial malformations are Treacher Collins syndrome and Acrofacial Dysostosis, Cincinnati type which have phenotypic overlap including deformities of the eyes, ears, and facial bones. Mutations in TCOF1, POLR1C or POLR1D may cause Treacher Collins syndrome while mutations in POLR1A may cause Acrofacial Dysostosis, Cincinnati type. TCOF1 encodes the nucleolar phosphoprotein Treacle, which functions in rRNA transcription and modification. Previous studies demonstrated that Tcof1 mutations in mice result in reduced ribosome biogenesis and increased neuroepithelial apoptosis. This diminishes the neural crest cell (NCC) progenitor population which contribute to the development of the cranial skeleton. In contrast, apart from being subunits of RNA Polymerases (RNAP) I and/or III, nothing is known about the function of POLR1A, POLR1C, and POLR1D during embryonic and craniofacial development. -

Supplemental Material.Pdf
Supplemental Material ZNF750 Interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 Chromatin Regulators to Repress Epidermal Progenitor Genes and Induce Differentiation Genes Lisa D. Boxer, Brook Barajas, Shiying Tao, Jiajing Zhang, and Paul Khavari Supplemental Inventory Figure S1. This figure supports Figure 1 and shows the Gene Ontology terms for ZNF750-bound but unaffected genes, the genomic enrichment of ZNF750 ChIP-seq peaks, and the percentage of peaks that contain the ZNF750 motif. Figure S2. This figure supports Figure 2 and shows gene expression changes with depletion of ZNF750-interacting proteins, confirms the knock-down of ZNF750-interacting proteins, and shows the quantification of Ki67 in ZNF750-interacting protein depleted organotypic tissue. Figure S3. This figure supports Figure 3 and shows the quantification of ZNF750 and interacting protein co-IPs, and IPs and Far western blots demonstrating competition between KLF4 and KDM1A for binding to ZNF750. Figure S4. This figure supports Figure 4 and shows the expression of ZNF750 mutant proteins and the effects of full-length or mutant ZNF750 on differentiation in organotypic tissue and on clonogenic growth. Figure S5. This figure supports Figure 5 and shows the effects of mutagenesis of ZNF750 and KLF4 motifs on reporter activity, and the changes in histone marks with depletion of ZNF750 and interacting proteins. Figure S6. This figure supports Figure 6 and shows the changes in mRNA and protein expression of ZNF750-interacting proteins during keratinocyte differentiation, and the expression of ZNF750-interacting proteins with ZNF750 depletion. Table S1. Supports Figure 1 and shows the genomic coordinates of ZNF750 ChIP-seq peaks. -

Regulation of Cell Fate Decisions in the Developing Forebrain by DLX
Regulation of cell fate decisions in the developing forebrain by DLX transcription factors by Mikaela Nevin A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science Medical Sciences- Medical Genetics University of Alberta © Mikaela Nevin, 2020 Abstract Introduction The Dlx1/Dlx2 double knockout (DKO) mouse exhibits major defects in forebrain development including a block in differentiation and migration of GABAergic interneurons from the subpallium to the cortex. In addition, the Dlx1/Dlx2 DKO mouse generates more oligodendrocyte progenitor cells (OPC) during forebrain development, and this occurs at the expense of interneuron generation. This phenotype suggests a role for DLX genes in mediating the neuronal-glial cell fate switch in the developing forebrain. Furthermore, expression of many oligodendroglial lineage genes is increased and occurs earlier in the DKO forebrain. As such, we hypothesized that DLX2 actively represses acquisition of oligodendroglial cell fate in a subset of forebrain neural progenitor cells by direct transcriptional repression of multiple genes required for oligodendroglial development. Here, I aimed to determine whether DLX2 represses expression of the oligodendroglial lineage genes Myt1 and Plp1 during forebrain development. In order to further characterise this gene regulatory network, I also investigated regulation of Plp1 by MYT1. I also investigated whether DLX2 may play a role in regulating progenitor cell fate in the pediatric brain tumour Diffuse Intrinsic Pontine Glioma (DIPG). Methods Chromatin immunoprecipitation with a polyclonal DLX2 antibody was performed on E13.5 mouse forebrain chromatin to determine whether DLX2 occupied candidate promoter regulatory regions of the Myt1 and Plp1 promoters, and with an MYT1 antibody on E14.5 and E18.5 forebrain to determine if MYT1 occupied candidate regulatory regions of the Plp1 promoter. -

The Tumor Suppressor HHEX Inhibits Axon Growth When Prematurely Expressed in Developing Central Nervous System Neurons
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by epublications@Marquette Marquette University e-Publications@Marquette Biological Sciences Faculty Research and Biological Sciences, Department of Publications 9-1-2015 The umorT Suppressor HHEX Inhibits Axon Growth when Prematurely Expressed in Developing Central Nervous System Neurons Matthew .T Simpson Marquette University Ishwariya Venkatesh Marquette University Ben L. Callif Marquette University Laura K. Thiel Marquette University Denise M. Coley Marquette University See next page for additional authors Accepted version. Molecular and Cellular Neuroscience, Vol 68 )September 2015): 272-283. DOI. © 2015 Elsevier Inc. Used with permission. NOTICE: this is the author’s version of a work that was accepted for publication in Molecular and Cellular Neuroscience. Changes resulting from the publishing process, such as peer review, editing, corrections, structural formatting, and other quality control mechanisms may not be reflected in this document. Changes may have been made to this work since it was submitted for publication. A definitive version was subsequently published in Molecular and Cellular Neuroscience, Vol 68 )September 2015): 272-283. DOI. Authors Matthew T. Simpson, Ishwariya Venkatesh, Ben L. Callif, Laura K. Thiel, Denise M. Coley, Kristen N. Winsor, Zimei Wang, Audra A. Kramer, Jessica K. Lerch, and Murray G. Blackmore This article is available at e-Publications@Marquette: https://epublications.marquette.edu/bio_fac/515 NOT THE PUBLISHED VERSION; this is the author’s final, peer-reviewed manuscript. The published version may be accessed by following the link in the citation at the bottom of the page. The Tumor Suppressor HHEX Inhibits Axon Growth When Prematurely Expressed in Developing Central Nervous System Neurons Matthew T. -

DLX Genes: Roles in Development and Cancer
cancers Review DLX Genes: Roles in Development and Cancer Yinfei Tan 1,* and Joseph R. Testa 1,2,* 1 Genomics Facility, Fox Chase Cancer Center, Philadelphia, PA 19111, USA 2 Cancer Signaling and Epigenetics Program, Fox Chase Cancer Center, Philadelphia, PA 19111, USA * Correspondence: [email protected] (Y.T.); [email protected] (J.R.T.) Simple Summary: DLX homeobox family genes encode transcription factors that have indispensable roles in embryonic and postnatal development. These genes are critically linked to the morphogene- sis of craniofacial structures, branchial arches, forebrain, and sensory organs. DLX genes are also involved in postnatal homeostasis, particularly hematopoiesis and, when dysregulated, oncogen- esis. DLX1/2, DLX3/4, and DLX5/6 exist as bigenes on different chromosomes, sharing intergenic enhancers between gene pairs, which allows orchestrated spatiotemporal expression. Genomic alterations of human DLX gene enhancers or coding sequences result in congenital disorders such as split-hand/foot malformation. Aberrant postnatal expression of DLX genes is associated with hematological malignancies, including leukemias and lymphomas. In several mouse models of T-cell lymphoma, Dlx5 has been shown to act as an oncogene by cooperating with activated Akt, Notch1/3, and/or Wnt to drive tumor formation. In humans, DLX5 is aberrantly expressed in lung and ovarian carcinomas and holds promise as a therapeutic target. Abstract: Homeobox genes control body patterning and cell-fate decisions during development. The homeobox genes consist of many families, only some of which have been investigated regarding a possible role in tumorigenesis. Dysregulation of HOX family genes have been widely implicated in cancer etiology.