The Platypus Is Not a Rodent: DNA Hybridization, Amniote Phylogeny and the Palimpsest Theory
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of the Patellar Sesamoid Bone in Mammals
A peer-reviewed version of this preprint was published in PeerJ on 21 March 2017. View the peer-reviewed version (peerj.com/articles/3103), which is the preferred citable publication unless you specifically need to cite this preprint. Samuels ME, Regnault S, Hutchinson JR. 2017. Evolution of the patellar sesamoid bone in mammals. PeerJ 5:e3103 https://doi.org/10.7717/peerj.3103 Evolution of the patellar sesamoid bone in mammals Mark E Samuels 1, 2 , Sophie Regnault 3 , John R Hutchinson Corresp. 3 1 Department of Medicine, University of Montreal, Montreal, Quebec, Canada 2 Centre de Recherche du CHU Ste-Justine, Montreal, Quebec, Canada 3 Structure & Motion Laboratory, Department of Comparative Biomedical Sciences, The Royal Veterinary College, Hatfield, Hertfordshire, United Kingdom Corresponding Author: John R Hutchinson Email address: [email protected] The patella is a sesamoid bone located in the major extensor tendon of the knee joint, in the hindlimb of many tetrapods. Although numerous aspects of knee morphology are ancient and conserved among most tetrapods, the evolutionary occurrence of an ossified patella is highly variable. Among extant (crown clade) groups it is found in most birds, most lizards, the monotreme mammals and almost all placental mammals, but it is absent in most marsupial mammals as well as many reptiles. Here we integrate data from the literature and first-hand studies of fossil and recent skeletal remains to reconstruct the evolution of the mammalian patella. We infer that bony patellae most likely evolved between four to six times in crown group Mammalia: in monotremes, in the extinct multituberculates, in one or more stem-mammal genera outside of therian or eutherian mammals, and up to three times in therian mammals. -

Altriciality and the Evolution of Toe Orientation in Birds
Evol Biol DOI 10.1007/s11692-015-9334-7 SYNTHESIS PAPER Altriciality and the Evolution of Toe Orientation in Birds 1 1 1 Joa˜o Francisco Botelho • Daniel Smith-Paredes • Alexander O. Vargas Received: 3 November 2014 / Accepted: 18 June 2015 Ó Springer Science+Business Media New York 2015 Abstract Specialized morphologies of bird feet have trees, to swim under and above the water surface, to hunt and evolved several times independently as different groups have fish, and to walk in the mud and over aquatic vegetation, become zygodactyl, semi-zygodactyl, heterodactyl, pam- among other abilities. Toe orientations in the foot can be prodactyl or syndactyl. Birds have also convergently described in six main types: Anisodactyl feet have digit II evolved similar modes of development, in a spectrum that (dII), digit III (dIII) and digit IV (dIV) pointing forward and goes from precocial to altricial. Using the new context pro- digit I (dI) pointing backward. From the basal anisodactyl vided by recent molecular phylogenies, we compared the condition four feet types have arisen by modifications in the evolution of foot morphology and modes of development orientation of digits. Zygodactyl feet have dI and dIV ori- among extant avian families. Variations in the arrangement ented backward and dII and dIII oriented forward, a condi- of toes with respect to the anisodactyl ancestral condition tion similar to heterodactyl feet, which have dI and dII have occurred only in altricial groups. Those groups repre- oriented backward and dIII and dIV oriented forward. Semi- sent four independent events of super-altriciality and many zygodactyl birds can assume a facultative zygodactyl or independent transformations of toe arrangements (at least almost zygodactyl orientation. -

Constraints on the Timescale of Animal Evolutionary History
Palaeontologia Electronica palaeo-electronica.org Constraints on the timescale of animal evolutionary history Michael J. Benton, Philip C.J. Donoghue, Robert J. Asher, Matt Friedman, Thomas J. Near, and Jakob Vinther ABSTRACT Dating the tree of life is a core endeavor in evolutionary biology. Rates of evolution are fundamental to nearly every evolutionary model and process. Rates need dates. There is much debate on the most appropriate and reasonable ways in which to date the tree of life, and recent work has highlighted some confusions and complexities that can be avoided. Whether phylogenetic trees are dated after they have been estab- lished, or as part of the process of tree finding, practitioners need to know which cali- brations to use. We emphasize the importance of identifying crown (not stem) fossils, levels of confidence in their attribution to the crown, current chronostratigraphic preci- sion, the primacy of the host geological formation and asymmetric confidence intervals. Here we present calibrations for 88 key nodes across the phylogeny of animals, rang- ing from the root of Metazoa to the last common ancestor of Homo sapiens. Close attention to detail is constantly required: for example, the classic bird-mammal date (base of crown Amniota) has often been given as 310-315 Ma; the 2014 international time scale indicates a minimum age of 318 Ma. Michael J. Benton. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Philip C.J. Donoghue. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Robert J. -

Predictors of Juvenile Survival in Birds
Ornithological Monographs Volume (2013), No. 78, 1–55 © The American Ornithologists’ Union, 2013. Printed in USA. PREDICTORS OF JUVENILE SURVIVAL IN BIRDS TERRI J. MANESS1,2,3 AND DAVID J. ANDERSON2 1School of Biological Sciences, Louisiana Tech University, Ruston, Louisiana 71272, USA; and 2Department of Biology, Wake Forest University, Winston-Salem, North Carolina 27106, USA ABSTRACT.—The survival probability of birds during the juvenile period, between the end of parental care and adulthood, is highly variable and has a major effect on population dynamics and parental fitness. As such, a large number of studies have attempted to evaluate potential predictors of juvenile survival in birds, especially predictors related to parental care. Lack’s hypothesis linking body reserves accumulated from parental care to the survival of naive juveniles has organized much of this research, but various other predictors have also been investigated and received some support. We reviewed the literature in this area and identified a variety of methodological problems that obscure interpretation of the body of results. Most studies adopted statistical techniques that missed the opportunities to (1) evaluate the relative importance of several predictors, (2) control the confounding effect of correlation among predictor variables, and (3) exploit the information content of collinearity by evaluating indirect (via correlation) as well as direct effects of potential predictors on juvenile survival. Ultimately, we concluded that too few reliable studies exist to allow robust evaluations of any hypothesis regarding juvenile survival in birds. We used path analysis to test potential predictors of juvenile survival of 2,631 offspring from seven annual cohorts of a seabird, the Nazca Booby (Sula granti). -

Early Cretaceous Amphilestid ('Triconodont') Mammals from Mongolia
Early Cretaceous amphilestid ('triconodont') mammals from Mongolia ZOFIAKIELAN-JAWOROWSKA and DEMBERLYIN DASHZEVEG Kielan-Jaworowską Z. &Daslueveg, D. 1998. Early Cretaceous amphilestid (.tricono- dont') mammals from Mongotia. - Acta Pal.aeontol.ogicaPolonica,43,3, 413438. Asmall collection of ?Aptianor ?Albian amphilestid('triconodont') mammals consisting of incomplete dentaries and maxillae with teeth, from the Khoboor localiĘ Guchin Us counĘ in Mongolia, is described. Grchinodon Troftmov' 1978 is regarded a junior subjective synonym of GobiconodonTroftmov, 1978. Heavier wear of the molariforms M3 andM4than of themore anteriorone-M2 in Gobiconodonborissiaki gives indirect evidence formolariformreplacement in this taxon. The interlocking mechanismbetween lower molariforms n Gobiconodon is of the pattern seen in Kuchneotherium and Ttnodon. The ińterlocking mechanism and the type of occlusion ally Amphilestidae with Kuehneotheriidae, from which they differ in having lower molariforms with main cusps aligned and the dentary-squamosal jaw joint (double jaw joint in Kuehneotheńdae). The main cusps in upper molariforms M3-M5 of Gobiconodon, however, show incipient tńangular arrangement. The paper gives some support to Mills' idea on the therian affinities of the Amphilestidae, although it cannot be excluded that the characters that unite the two groups developed in parallel. Because of scanty material and arnbiguĘ we assign the Amphilestidae to order incertae sedis. Key words : Mammali4 .triconodonts', Amphilestidae, Kuehneotheriidae, Early Cretaceous, Mongolia. Zofia Kiel,an-Jaworowska [zkielnn@twarda,pan.pl], InsĘtut Paleobiologii PAN, ul. Twarda 5 I /5 5, PL-00-8 I 8 Warszawa, Poland. DemberĘin Dash7eveg, Geological Institute, Mongolian Academy of Sciences, Ulan Bator, Mongolia. Introduction Beliajeva et al. (1974) reportedthe discovery of Early Cretaceous mammals at the Khoboor locality (referred to also sometimes as Khovboor), in the Guchin Us Soinon (County), Gobi Desert, Mongolia. -

Mesozoic: the Dark Age for Mammals!
Ed’s Simplified History of the Mammals Note progression from Pelycosaurs (1) to Therapsids and Cynodonts (2) in Triassic. Stem mammals appeared in Late Triassic and Early Jurassic (3). Relationships among the Middle Jurassic forms (4) are controversial (see handout). Therian clade, identified by the tribosphenic molar (5), emerged at the end of the Jurassic, Early Cretaceous. A slightly more detailed version… in case you like something that looks more slick From Pough et al. 2009. Vertebrate Life, 8th ed. Pelycosaurs Dominated the late Permian, gave rise to therapsids Therapsids Rapid radiation in late Permian, around 270 MYA Still “mammal-like reptiles” The mass extinction at the end of the Permian was the greatest loss of diversity ever with >80% of all known genera and about 90% of all species going extinct, both terrestrial and marine. Cynodonts Late Permian to mid Triassic Last remaining group of therapsids, survived mass extinction at the end of the Permian. Persisted well Only 1 lineage of into Triassic and developed cynodonts survived many features associated through the late Triassic, with mammals. and this group became ancestors of mammals. Mesozoic: the Dark Age for Mammals! multituberculate Morganucodon, one of the earliest mammals (What else was happening in the Late Triassic and Jurassic Hadrocodium that may have contributed to mammals becoming small and Most were very small with nocturnal?) conservative morphology ...but new fossil finds indicate more diversity than we thought Repenomanus Still, largest known mammal during Mesozic Most were shrew to is no larger than a mouse sized, for 125 woodchuck million years! Some Mesozoic events and mammals you should know 1. -

Paleobiology of Jurassic Mammals. George Gaylord Simpson
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Palaeobiologica Jahr/Year: 1933 Band/Volume: 5 Autor(en)/Author(s): Simpson George Gaylord Artikel/Article: Paleobiology of Jurassic Mammals. 127-158 P aleobiology o f J u r a ss ic M a m m a l s . By George Gaylord S im pson (New York). With 6 figures. Pag. Introduction . 127 Occlusion, food habits, and dental evolution 128 Multituberculata 133 Triconodonta 133 Symmetrodonta 137 Pantotheria 139 Environments and ecology 145 Stonesfield 146 Purbeck 147 Morrison 150 Conclusions 155 References 158 Introduction. Of the numerous problems relating to the rise of the Mammalia, some of the most important are paleobiological in nature. True understanding of the early history of mammals involves not only knowledge of their morphology, but also some conception of the conditions under which they lived, of their habits, and of their place in the Mesozoic faunas. Such a study involves three related lines of inquiry: first, the habits of the mammals themselves, so far as these can be inferred from their imperfect remains; second, the nature of the environ ment in which they lived; and third, their ecological relationships to the accompanying biota. This is almost a virgin field for research. The habits of a single group, the Multituberculata, have been dis cussed at some length by various students, and will be only briefly mentioned here, but the three other orders have not been studied from a paleobiological point of view. The four Jurassic orders, Multituberculata, Triconodonta, Symmetrodonta, and Pantotheria, PALAEOBIOLOGICA, Band V. -

Mammalian Organogenesis in Deep Time: Tools for Teaching and Outreach Marcelo R
Sánchez‑Villagra and Werneburg Evo Edu Outreach (2016) 9:11 DOI 10.1186/s12052-016-0062-y REVIEW Open Access Mammalian organogenesis in deep time: tools for teaching and outreach Marcelo R. Sánchez‑Villagra1 and Ingmar Werneburg1,2,3,4* Abstract Mammals constitute a rich subject of study on evolution and development and provide model organisms for experi‑ mental investigations. They can serve to illustrate how ontogeny and phylogeny can be studied together and how the reconstruction of ancestors of our own evolutionary lineage can be approached. Likewise, mammals can be used to promote ’tree thinking’ and can provide an organismal appreciation of evolutionary changes. This subject is suitable for the classroom and to the public at large given the interest and familiarity of people with mammals and their closest relatives. We present a simple exercise in which embryonic development is presented as a transforma‑ tive process that can be observed, compared, and analyzed. In addition, we provide and discuss a freely available animation on organogenesis and life history evolution in mammals. An evolutionary tree can be the best tool to order and understand those transformations for different species. A simple exercise introduces the subject of changes in developmental timing or heterochrony and its importance in evolution. The developmental perspective is relevant in teaching and outreach efforts for the understanding of evolutionary theory today. Keywords: Development, Ontogeny, Embryology, Phylogeny, Heterochrony, Recapitulation, Placentalia, Human Background (Gilbert 2013), followed by the growth process. In pla- Mammals are a diverse group in which to examine devel- cental mammals, organogenesis takes place mostly in the opment and evolution, and besides the mouse and the rat uterus, whereas in monotremes and marsupials a very used in biomedical research, provide subjects based on immature hatchling or newborn, respectively, develops which experimental (Harjunmaa et al. -
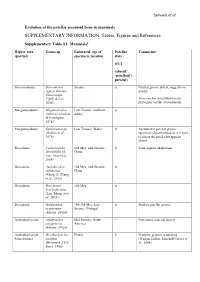
SUPPLEMENTARY INFORMATION: Tables, Figures and References
Samuels et al. Evolution of the patellar sesamoid bone in mammals SUPPLEMENTARY INFORMATION: Tables, Figures and References Supplementary Table S1: Mammals$ Higher taxa Genus sp. Estimated. age of Patellar Comments# (partial) specimen, location state 0/1/2 (absent/ ‘patelloid’/ present) Sinoconodonta Sinoconodon Jurassic 0 Patellar groove absent, suggests no rigneyi (Kielan- patella Jaworowska, Cifelli & Luo, Sinoconodon is included on our 2004) phylogeny within tritylodontids. Morganucodonta Megazostrodon Late Triassic, southern 0 rudnerae (Jenkins Africa & Parrington, 1976) Morganucodonta Eozostrodon sp. Late Triassic, Wales 0 Asymmetric patellar groove, (Jenkins et al., specimens disarticulated so it is hard 1976) to assess the patella but appears absent Docodonta Castorocauda 164 Mya, mid-Jurassic, 0 Semi-aquatic adaptations lutrasimilis (Ji, China Luo, Yuan et al., 2006) Docodonta Agilodocodon 164 Mya, mid-Jurassic, 0 scansorius China (Meng, Ji, Zhang et al., 2015) Docodonta Docofossor 160 Mya 0 brachydactylus (Luo, Meng, Ji et al., 2015) Docodonta Haldanodon 150-155 Mya, Late 0 Shallow patellar groove exspectatus Jurassic, Portugal (Martin, 2005b) Australosphenida Asfaltomylos Mid-Jurassic, South ? Postcranial material absent patagonicus America (Martin, 2005a) Australosphenida Ornithorhynchus Extant 2 Platypus, genome sequenced Monotremata anatinus (Warren, Hillier, Marshall Graves et (Herzmark, 1938; al., 2008) Rowe, 1988) Samuels et al. Australosphenida Tachyglossus + Extant 2 Echidnas Monotremata Zaglossus spp. (Herzmark, 1938; Rowe, 1988) Mammaliaformes Fruitafossor 150 Mya, Late Jurassic, 0 Phylogenetic status uncertain indet. windscheffeli (Luo Colorado & Wible, 2005) Mammaliaformes Volaticotherium Late Jurassic/Early ? Hindlimb material incomplete indet. antiquus (Meng, Cretaceous Hu, Wang et al., 2006) Eutriconodonta Jeholodens 120-125 Mya, Early 0 Poorly developed patellar groove jenkinsi (Ji, Luo Cretaceous, China & Ji, 1999) Eutriconodonta Gobiconodon spp. -

Martin Pj Edwardes an Anthropological Perspective
The Origins of Self explores the role that selfhood plays in defining human society, and each human individual in that society. It considers the genetic and cultural origins of self, the role that self plays in socialisation and language, and the types of self we generate in our individual journeys to and through adulthood. Edwardes argues that other awareness is a relatively early evolutionary development, present throughout the primate clade and perhaps beyond, but self-awareness is a product of the sharing of social models, something only humans appear to do. The self of which we are aware is not something innate within us, it is a model of our self produced as a response to the models of us offered to us by other people. Edwardes proposes that human construction of selfhood involves seven different types of self. All but one of them are internally generated models, and the only non-model, the actual self, is completely hidden from conscious awareness. We rely on others to tell us about our self, and even to let us know we are a self. Developed in relation to a range of subject areas – linguistics, anthropology, genomics and cognition, as well as socio-cultural theory – The Origins of Self is of particular interest to MARTIN P. J. EDWARDES students and researchers studying the origins of language, human origins in general, and the cognitive differences between human and other animal psychologies. Martin P. J. Edwardes is a visiting lecturer at King’s College London. He is currently teaching modules on Language Origins and Language Construction. -

Paleontology Lab: Mammalian Diversity
Paleontology Lab: Mammalian diversity Objectives: • Be able to recognize a mammal. • Be able to describe mammalian traits and evolutionary adaptations. • Be able to identify the major orders of mammals. • Be able to identify the characteristics of primates. Materials needed for this lab : Preserved specimens and skeletons of various mammals including primates Human brain and sheep brain for comparison Class Mammalia: Mammals are warm-blooded vertebrates with a fully divided (four-chambered) heart that nourish their young with milk. Most mammals are furry, but some marine mammals have a thick layer of subcutaneous fat instead. Living mammals can be divided into a few egg- laying species (the Monotremes, including the platypus and echidna), the pouched mammals (Marsupials, including opossums, kangaroos, koalas, and others), and the placental mammals ( Eutheria ). If we include fossil lineages, we might more appropriately divide mammals into Protheria and Theria. Subclass Prototheria: These are mammals whose molar teeth have cusps in antero-posterior rows and whose petrosal bone is exposed within the eye socket. Most of these mammals are extinct. Extinct prototherians include the Triconodonta, Docodonta, and Multituberculata. Order Monotremata: Egg-laying mammals, including only the platypus and the echidna ("spiny anteater"), both now confined to Australia and New Guinea. Examine any monotreme specimens that may be available. Subclass Theria: Therian mammals, with molar teeth whose molar cusps were originally arranged in opposing triangles with shear surfaces between them, the so-called tribosphenic tooth pattern. The petrosal bone is never exposed inside the eye socket (orbit). Therian mammals are divided into infraclasses Pantotheria, Metatheria, and Theria. Infraclass Pantotheria includes the extinct orders Symmetrodonta and Eupantotheria. -

Mammalian Fauna of the Late Jurassic Guimarota Ecosystem
Asociación Paleontológica Argentina. Publicación Especial 7 ISSN 0328-347X VII International Symposium on Mesozoic Terrestrial Ecosystems: 123-126. Buenos Aires, 30-6-2001 MaMMalian fauna of the Late Jurassic Guimarota ecoSyStem Thomas MARTIN1 Abstract. The Late Jurassic (Kirnmeridgian) Guimarota loca lit y near Leiria in west-central Portugal has yielded more than 800 mammalian dentaries and partial skulls representing the largest sample of Late Jurassic mammals in the world. However, despite the enormous number of specimens collected, the mam- malian fauna appears somewhat depauperate. So far, only Multituberculata (several genera of Paulchoffatiidae; 28% of total number), Docodonta (Halda nadan exspectaius Kühne and Krusat; 24%), and Holotheria (48%) represented by Paurodontidae (Henkelatherium guimaratae Krebs and Drescheratherium acutum Krebs) and Dryolestidae (Dryalestes leiriensis Martín, Krebsoiherium Iusiianicum Martin, and Guimaroiodus inflatus Martin) have been detected. "Triconodonta" and Syrnmetrodonta, which are well represented at other Late Jurassic localities (e.g., Morrison Forrnation), are missing. To recover the rare mammalian taxa of the Guimarota ecosystem, a project was initiated to study the nearly 7000 isolated mammalian teeth that had been obtained by screenwashing. With a single exception, the same taxa are represented. Among the thousands of isolated teeth, however, 25 lower and 23 upper premolars and mo- lars of a tiny primitive Zatheria have been found, of which the lower molars closely resemble the "Porto Pinheiro molar". After sorting the isolated teeth, the mammalian fauna of the Guimarota ecosystem prob- ably is completely recorded. Apparently, the coastal swamp in the Lusitanian graben where the lignite formed represented a stressed environment not appropriate for some marnmalian groups.