1 Complete List of Variables Collected
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LRP2 Is Associated with Plasma Lipid Levels 311 Original Article
310 Journal of Atherosclerosis and Thrombosis Vol.14, No.6 LRP2 is Associated with Plasma Lipid Levels 311 Original Article Genetic Association of Low-Density Lipoprotein Receptor-Related Protein 2 (LRP2) with Plasma Lipid Levels Akiko Mii1, 2, Toshiaki Nakajima2, Yuko Fujita1, Yasuhiko Iino1, Kouhei Kamimura3, Hideaki Bujo4, Yasushi Saito5, Mitsuru Emi2, and Yasuo Katayama1 1Department of Internal Medicine, Divisions of Neurology, Nephrology, and Rheumatology, Nippon Medical School, Tokyo, Japan. 2Department of Molecular Biology-Institute of Gerontology, Nippon Medical School, Kawasaki, Japan. 3Awa Medical Association Hospital, Chiba, Japan. 4Department of Genome Research and Clinical Application, Graduate School of Medicine, Chiba University, Chiba, Japan. 5Department of Clinical Cell Biology, Graduate School of Medicine, Chiba University, Chiba, Japan. Aim: Not all genetic factors predisposing phenotypic features of dyslipidemia have been identified. We studied the association between the low density lipoprotein-related protein 2 gene (LRP2) and levels of plasma total cholesterol (T-Cho) and LDL-cholesterol (LDL-C) among 352 adults in Japan. Methods: Subjects were obtained from among participants in a cohort study that was carried out with health-check screening in an area of east-central Japan. We selected 352 individuals whose LDL-C levels were higher than 140 mg/dL from the initially screened 22,228 people. We assessed the relation between plasma cholesterol levels and single-nucleotide polymorphisms (SNPs) in the LRP2 gene. Results: -

Potential Microrna-Related Targets in Clearance Pathways of Amyloid-Β
Madadi et al. Cell Biosci (2019) 9:91 https://doi.org/10.1186/s13578-019-0354-3 Cell & Bioscience REVIEW Open Access Potential microRNA-related targets in clearance pathways of amyloid-β: novel therapeutic approach for the treatment of Alzheimer’s disease Soheil Madadi1, Heidi Schwarzenbach2, Massoud Saidijam3, Reza Mahjub4 and Meysam Soleimani1* Abstract Imbalance between amyloid-beta (Aβ) peptide synthesis and clearance results in Aβ deregulation. Failure to clear these peptides appears to cause the development of Alzheimer’s disease (AD). In recent years, microRNAs have become established key regulators of biological processes that relate among others to the development and progres- sion of neurodegenerative diseases, such as AD. This review article gives an overview on microRNAs that are involved in the Aβ cascade and discusses their inhibitory impact on their target mRNAs whose products participate in Aβ clear- ance. Understanding of the mechanism of microRNA in the associated signal pathways could identify novel therapeu- tic targets for the treatment of AD. Keywords: Ubiquitin–proteasome system, Autophagy, Aβ-degrading proteases, BBB transporters, Phagocytosis, Heat shock proteins, microRNAs Introduction stage, APP is cleaved to non-toxic proteins by α-secretase Alzheimer’s disease (AD)—the most common form of [6]. Aβ has two major forms: Aβ40 and Aβ42, which are dementia—is a devastating diagnosis that accounts for 40 and 42 amino acid-long fragments, respectively. Since 93,541 deaths in the United States in 2014 [1]. Clinical Aβ42 is more hydrophobic than Aβ40, it is more prone to manifestation of AD is often a loss of memory and cog- aggregate and scafold for oligomeric and fbrillar forms nitive skills. -

Human Megalin/LRP2 Antibody
Human Megalin/LRP2 Antibody Monoclonal Mouse IgG1 Clone # 545606 Catalog Number: MAB9578 DESCRIPTION Species Reactivity Human Specificity Detects human Megalin/LRP2 in direct ELISAs. Source Monoclonal Mouse IgG1 Clone # 545606 Purification Protein A or G purified from ascites Immunogen Mouse myeloma cell line NS0derived recombinant human Megalin/LRP2 Pro3510Lys3964 Accession # P98164 Formulation Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. See Certificate of Analysis for details. *Small pack size (SP) is supplied either lyophilized or as a 0.2 μm filtered solution in PBS. APPLICATIONS Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website. Recommended Sample Concentration Flow Cytometry 0.25 µg/106 cells See Below Immunohistochemistry 525 µg/mL See Below CyTOFready Ready to be labeled using established conjugation methods. No BSA or other carrier proteins that could interfere with conjugation. DATA Flow Cytometry Immunohistochemistry Detection of Megalin/LRP2 in CaCo2 Megalin/LRP2 in Human Kidney. Human Cell Line by Flow Cytometry. Megalin/LRP2 was detected in immersion CaCo2 human cell line was stained with fixed paraffinembedded sections of human Mouse AntiHuman Megalin/LRP2 kidney using Mouse AntiHuman Monoclonal Antibody (Catalog # MAB9578, Megalin/LRP2 Monoclonal Antibody (Catalog filled histogram) or isotype control antibody # MAB9578) at 5 µg/mL for 1 hour at room (Catalog # MAB002, open histogram), temperature followed by incubation with the followed by Phycoerythrinconjugated Anti AntiMouse IgG VisUCyte™ HRP Polymer Mouse IgG Secondary Antibody (Catalog # Antibody (Catalog # VC001). -

Clusterin and LRP2 Are Critical Components of the Hypothalamic Feeding Regulatory Pathway
ARTICLE Received 21 Sep 2012 | Accepted 16 Apr 2013 | Published 14 May 2013 DOI: 10.1038/ncomms2896 Clusterin and LRP2 are critical components of the hypothalamic feeding regulatory pathway So Young Gil1, Byung-Soo Youn2, Kyunghee Byun3,4, Hu Huang5, Churl Namkoong1, Pil-Geum Jang1, Joo-Yong Lee1, Young-Hwan Jo6, Gil Myoung Kang1, Hyun-Kyong Kim1, Mi-Seon Shin7, Claus U. Pietrzik8, Bonghee Lee3,4, Young-Bum Kim3,5 & Min-Seon Kim1,7 Hypothalamic feeding circuits are essential for the maintenance of energy balance. There have been intensive efforts to discover new biological molecules involved in these pathways. Here we report that central administration of clusterin, also called apolipoprotein J, causes anorexia, weight loss and activation of hypothalamic signal transduction-activated transcript-3 in mice. In contrast, inhibition of hypothalamic clusterin action results in increased food intake and body weight, leading to adiposity. These effects are likely mediated through the mutual actions of the low-density lipoprotein receptor-related protein-2, a potential receptor for clusterin, and the long-form leptin receptor. In response to clusterin, the low-density lipoprotein receptor-related protein-2 binding to long-form leptin receptor is greatly enhanced in cultured neuronal cells. Furthermore, long-form leptin receptor deficiency or hypothalamic low-density lipoprotein receptor-related protein-2 suppression in mice leads to impaired hypothalamic clusterin signalling and actions. Our study identifies the hypotha- lamic clusterin–low-density lipoprotein receptor-related protein-2 axis as a novel anorexigenic signalling pathway that is tightly coupled with long-form leptin receptor-mediated signalling. 1 Asan Institute for Life Science, University of Ulsan College of Medicine, Seoul 138-736, Korea. -

Coexpression of ATP-Binding Cassette Proteins ABCG5 and ABCG8 Permits Their Transport to the Apical Surface
Coexpression of ATP-binding cassette proteins ABCG5 and ABCG8 permits their transport to the apical surface Gregory A. Graf, … , Jonathan C. Cohen, Helen H. Hobbs J Clin Invest. 2002;110(5):659-669. https://doi.org/10.1172/JCI16000. Article Cardiology Mutations in either ATP-binding cassette (ABC) G5 or ABCG8 cause sitosterolemia, an autosomal recessive disorder of sterol trafficking. To determine the site of action of ABCG5 and ABCG8, we expressed recombinant, epitope-tagged mouse ABCG5 and ABCG8 in cultured cells. Both ABCG5 and ABCG8 underwent N-linked glycosylation. When either protein was expressed individually in cells, the N-linked sugars remained sensitive to Endoglycosidase H (Endo H). When ABCG5 and ABCG8 were coexpressed, the attached sugars were Endo H–resistant and neuraminidase-sensitive, indicating that the proteins were transported to the trans-Golgi complex. The mature, glycosylated forms of ABCG5 and ABCG8 coimmunoprecipitated, consistent with heterodimerization of these two proteins. The Endo H–sensitive forms of ABCG5 and ABCG8 were confined to the endoplasmic reticulum (ER), whereas the mature forms were present in non-ER fractions in cultured hepatocytes. Immunoelectron microscopy revealed ABCG5 and ABCG8 on the plasma membrane of these cells. In polarized WIF-B cells, recombinant ABCG5 localized to the apical (canalicular) membrane when coexpressed with ABCG8, but not when expressed alone. To our knowledge this is the first direct demonstration that trafficking of an ABC half-transporter to the cell surface requires the presence of its dimerization partner. Find the latest version: https://jci.me/16000/pdf Coexpression of ATP-binding cassette See the related Commentary beginning on page 605. -

Plasma Proteomic Analysis Reveals Altered Protein Abundances In
Lygirou et al. J Transl Med (2018) 16:104 https://doi.org/10.1186/s12967-018-1476-9 Journal of Translational Medicine RESEARCH Open Access Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease Vasiliki Lygirou1 , Agnieszka Latosinska2, Manousos Makridakis1, William Mullen3, Christian Delles3, Joost P. Schanstra4,5, Jerome Zoidakis1, Burkert Pieske6, Harald Mischak2 and Antonia Vlahou1* Abstract Background: Cardiovascular disease (CVD) describes the pathological conditions of the heart and blood vessels. Despite the large number of studies on CVD and its etiology, its key modulators remain largely unknown. To this end, we performed a comprehensive proteomic analysis of blood plasma, with the scope to identify disease-associated changes after placing them in the context of existing knowledge, and generate a well characterized dataset for fur- ther use in CVD multi-omics integrative analysis. Methods: LC–MS/MS was employed to analyze plasma from 32 subjects (19 cases of various CVD phenotypes and 13 controls) in two steps: discovery (13 cases and 8 controls) and test (6 cases and 5 controls) set analysis. Follow- ing label-free quantifcation, the detected proteins were correlated to existing plasma proteomics datasets (plasma proteome database; PPD) and functionally annotated (Cytoscape, Ingenuity Pathway Analysis). Diferential expression was defned based on identifcation confdence ( 2 peptides per protein), statistical signifcance (Mann–Whitney p value 0.05) and a minimum of twofold change.≥ ≤ Results: Peptides detected in at least 50% of samples per group were considered, resulting in a total of 3796 identi- fed proteins (838 proteins based on 2 peptides). Pathway annotation confrmed the functional relevance of the fndings (representation of complement≥ cascade, fbrin clot formation, platelet degranulation, etc.). -

Cardiovascular Disease Products
Cardiovascular Disease Products For more information, visit: www.bosterbio.com Cardiovascular Disease Research Cardiovascular disease is the leading cause of death in developed nations. Boster Bio aims to supply researchers with high-quality antibodies and ELISA kits so they can make new discoveries and help save lives. In this catalogue you will find a comprehensive list of high-affinity Boster antibodies and high sensitivity Boster ELISA kits targeted at proteins associated with cardiovascular disease. Boster: The Fastest Growing About Bosterbio Antibody Company In 2015 Boster is an antibody manufacturer founded in 1993 by histologist Steven Xia. Over the past two decades, Boster and its products have been cited in over 20,000 publications and counting. The firm specializes in developing antibodies and ELISA kits that feature high affinity, Boster Bio received the CitaAb award for high specificity at affordable the greatest increase in number of prices. citations during 2015 than any other antibody manufacturer. Table of Contents Boster Cardiovascular Disease Related Antibodies…………..………..... 2 Boster Cardiovascular Disease Related ELISA Kits……………………..…. 9 1 High Affinity Boster Antibodies Boster supplies only the highest quality antibodies. Our high-affinity polyclonal and monoclonal antibodies are thoroughly validated by Western Blotting, Immunohistochemistry and ELISA. This is our comprehensive catalog of our antibody products related to cardiovascular disease, sorted in alphabetical order by target gene name. Catalog No Product Name -
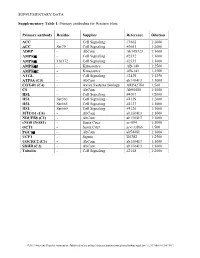
Supplementary Table 1
SUPPLEMENTARY DATA Supplementary Table 1. Primary antibodies for Western blots. Primary antibody Residue Supplier Reference Dilution ACC - Cell Signaling #3662 1:2000 ACC Ser79 Cell Signaling #3661 1:2000 ADRP - AbCam Ab108323 1:1000 AMPKα - Cell Signaling #2532 1:1000 AMPKα Thr172 Cell Signaling #2535 1:1000 AMPKα1 - Kinasource AB-140 1:2500 AMPKα2 - Kinasource AB-141 1:2500 ATGL - Cell Signaling #2439 1:1250 ATP5A (C5) - AbCam ab110413 1:1000 COX4l1 (C4) - Aviva Systems Biology ARP42784 1:500 CS - AbCam Ab96600 1:1000 HSL - Cell Signaling #4107 1:2000 HSL Ser563 Cell Signaling #4139 1:2000 HSL Ser565 Cell Signaling #4137 1:1000 HSL Ser660 Cell Signaling #4126 1:1000 MTCO1 (C4) - AbCam ab110413 1:1000 NDUFB8 (C1) - AbCam ab110413 1:1000 eNOS (NOS3) - Santa Cruz sc-654 1:1000 OCT1 - Santa Cruz sc-133866 1:500 PGC1α - AbCam ab54481 1:1000 UCP1 - Sigma U6382 1:2500 UQCRC2 (C3) - AbCam ab110413 1:1000 SDHB (C2) - AbCam ab110413 1:1000 Tubulin - Cell Signaling #2148 1:2000 ©2013 American Diabetes Association. Published online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-0194/-/DC1 SUPPLEMENTARY DATA Supplementary Table 2. Primer sequences for qRT-PCR. Gene Accession number Forward primer Reverse primer Abca1 NM_013454.3 CCCAGAGCAAAAAGCGACTC GGTCATCATCACTTTGGTCCTTG Abcg5 NM_031884 TGTCCTACAGCGTCAGCAACC GGCCACTCTCGATGTACAAGG Abcg8 NM_026180 TCCTGTGAGCTGGGCATCCGA CCCGCAGCCTGAGCTCCCTAT Acaca NM_133360.2 CAGCTGGTGCAGAGGTACCG TCTACTCGCAGGTACTGCCG Acacb NM_133904.2 GCGCCTACTATGAGGCCCAGCA ACAAACTCGGCTGGGGACGC Acly NM_134037.2 -

A Case of Ezetimibe-Effective Hypercholesterolemia with a Novel Heterozygous Variant in ABCG5
2020, 67 (11), 1099-1105 Original A case of ezetimibe-effective hypercholesterolemia with a novel heterozygous variant in ABCG5 Yujiro Nakano1), Chikara Komiya1), Hitomi Shimizu2), 3), Hiroyuki Mishima3), Kumiko Shiba1), Kazutaka Tsujimoto1), Kenji Ikeda1), Kenichi Kashimada4), Sumito Dateki2), Koh-ichiro Yoshiura3), Yoshihiro Ogawa5) and Tetsuya Yamada1) 1) Department of Molecular Endocrinology and Metabolism, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo 113-8519, Japan 2) Department of Pediatrics, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaski 852-8501, Japan 3) Department of Human Genetics, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaki 852-8501, Japan 4) Department of Pediatrics and Developmental Biology, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, Tokyo 113-8519, Japan 5) Department of Medicine and Bioregulatory Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan Abstract. Sitosterolemia is caused by homozygous or compound heterozygous gene mutations in either ATP-binding cassette subfamily G member 5 (ABCG5) or 8 (ABCG8). Since ABCG5 and ABCG8 play pivotal roles in the excretion of neutral sterols into feces and bile, patients with sitosterolemia present elevated levels of serum plant sterols and in some cases also hypercholesterolemia. A 48-year-old woman was referred to our hospital for hypercholesterolemia. She had been misdiagnosed with familial hypercholesterolemia at the age of 20 and her serum low-density lipoprotein cholesterol (LDL-C) levels had remained about 200–300 mg/dL at the former clinic. Although the treatment of hydroxymethylglutaryl-CoA (HMG-CoA) reductase inhibitors was ineffective, her serum LDL-C levels were normalized by ezetimibe, a cholesterol transporter inhibitor. -

Common Genetic Variations Involved in the Inter-Individual Variability Of
nutrients Review Common Genetic Variations Involved in the Inter-Individual Variability of Circulating Cholesterol Concentrations in Response to Diets: A Narrative Review of Recent Evidence Mohammad M. H. Abdullah 1 , Itzel Vazquez-Vidal 2, David J. Baer 3, James D. House 4 , Peter J. H. Jones 5 and Charles Desmarchelier 6,* 1 Department of Food Science and Nutrition, Kuwait University, Kuwait City 10002, Kuwait; [email protected] 2 Richardson Centre for Functional Foods & Nutraceuticals, University of Manitoba, Winnipeg, MB R3T 6C5, Canada; [email protected] 3 United States Department of Agriculture, Agricultural Research Service, Beltsville, MD 20705, USA; [email protected] 4 Department of Food and Human Nutritional Sciences, University of Manitoba, Winnipeg, MB R3T 2N2, Canada; [email protected] 5 Nutritional Fundamentals for Health, Vaudreuil-Dorion, QC J7V 5V5, Canada; [email protected] 6 Aix Marseille University, INRAE, INSERM, C2VN, 13005 Marseille, France * Correspondence: [email protected] Abstract: The number of nutrigenetic studies dedicated to the identification of single nucleotide Citation: Abdullah, M.M.H.; polymorphisms (SNPs) modulating blood lipid profiles in response to dietary interventions has Vazquez-Vidal, I.; Baer, D.J.; House, increased considerably over the last decade. However, the robustness of the evidence-based sci- J.D.; Jones, P.J.H.; Desmarchelier, C. ence supporting the area remains to be evaluated. The objective of this review was to present Common Genetic Variations Involved recent findings concerning the effects of interactions between SNPs in genes involved in cholesterol in the Inter-Individual Variability of metabolism and transport, and dietary intakes or interventions on circulating cholesterol concen- Circulating Cholesterol trations, which are causally involved in cardiovascular diseases and established biomarkers of Concentrations in Response to Diets: cardiovascular health. -

A New Role for LRP2 in Forebrain Development
A new role for LRP2 in forebrain development DISSERTATION zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) im Fach Biologie eingereicht an der Mathematisch-Naturwissenschaftlichen Fakult¨atI Humboldt-Universit¨atzu Berlin von Herr Dipl.-Biol. (technisch orientiert) Uwe Anzenberger geboren am 22.03.1976 in Singen a. Htwl. Pr¨asident der Humboldt-Universit¨atzu Berlin: Prof. Dr. Christoph Markschies Dekan der Mathematisch-Naturwissenschaftlichen Fakult¨atI: Prof. Thomas Buckhout, Phd Gutachter: 1. Prof. Dr. Thomas Willnow 2. Prof. Dr. Michael Bader 3. Prof. Dr. Wolfgang Lockau Tag der m¨undlichen Pr¨ufung: 22. September 2006 Abstract LRP2 is a member of the low-density lipoprotein receptor gene family that is mainly expressed in the yolk sac and in the neuroepithelium of the early embryo. Deficiency for this 600 kDa protein in mice results in holoprosencephaly, indicating an important yet unknown role for LRP2 in forebrain development. In this study, mice with a complete or a conditional loss of lrp2 function were used to further elucidate the consequences of the lack of LRP2 expression. This study shows that the presence of LRP2 in the neuroepithelium but not in the yolk sac is crucial for early forebrain development. Lack of the receptor resulted in an increase of Bone morphogenic protein (Bmp) 4 signaling in the rostral telencephalon at E9.5. As a consequence, sonic hedgehog (shh) expression at E10.5 was lost completely in a ventral region of the telencephalon termed anterior entopeduncular area (AEP). The absence of Shh activity in this area subsequently led to the loss of ventrally induced oligodendroglial and interneuronal cell populations in lrp2 deficient mice. -

University of Groningen Maternal-Fetal Cholesterol Transport in The
University of Groningen Maternal-fetal cholesterol transport in the second half ofmouse pregnancy does not involve LDL receptor-related protein 2 Zwier, Mathijs V; Baardman, Maria E; van Dijk, Theo H.; Jurdzinski, Angelika; Wisse, Lambertus J; Bloks, Vincent; Berger, Rolf M. F. ; DeRuiter, Marco C; Groen, Albert; Plosch, Torsten Published in: Acta physiologica DOI: 10.1111/apha.12845 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Final author's version (accepted by publisher, after peer review) Publication date: 2017 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Zwier, M. V., Baardman, M. E., van Dijk, T. H., Jurdzinski, A., Wisse, L. J., Bloks, V. W., ... Plösch, T. (2017). Maternal-fetal cholesterol transport in the second half ofmouse pregnancy does not involve LDL receptor-related protein 2. Acta physiologica, 220(4), 471-485. DOI: 10.1111/apha.12845 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum.