Uniprot - the Universal Protein Resource
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Uniprot at EMBL-EBI's Role in CTTV
Barbara P. Palka, Daniel Gonzalez, Edd Turner, Xavier Watkins, Maria J. Martin, Claire O’Donovan European Bioinformatics Institute (EMBL-EBI), European Molecular Biology Laboratory, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK UniProt at EMBL-EBI’s role in CTTV: contributing to improved disease knowledge Introduction The mission of UniProt is to provide the scientific community with a The Centre for Therapeutic Target Validation (CTTV) comprehensive, high quality and freely accessible resource of launched in Dec 2015 a new web platform for life- protein sequence and functional information. science researchers that helps them identify The UniProt Knowledgebase (UniProtKB) is the central hub for the collection of therapeutic targets for new and repurposed medicines. functional information on proteins, with accurate, consistent and rich CTTV is a public-private initiative to generate evidence on the annotation. As much annotation information as possible is added to each validity of therapeutic targets based on genome-scale experiments UniProtKB record and this includes widely accepted biological ontologies, and analysis. CTTV is working to create an R&D framework that classifications and cross-references, and clear indications of the quality of applies to a wide range of human diseases, and is committed to annotation in the form of evidence attribution of experimental and sharing its data openly with the scientific community. CTTV brings computational data. together expertise from four complementary institutions: GSK, Biogen, EMBL-EBI and Wellcome Trust Sanger Institute. UniProt’s disease expert curation Q5VWK5 (IL23R_HUMAN) This section provides information on the disease(s) associated with genetic variations in a given protein. The information is extracted from the scientific literature and diseases that are also described in the OMIM database are represented with a controlled vocabulary. -

Gene Discovery and Annotation Using LCM-454 Transcriptome Sequencing Scott J
Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Methods Gene discovery and annotation using LCM-454 transcriptome sequencing Scott J. Emrich,1,2,6 W. Brad Barbazuk,3,6 Li Li,4 and Patrick S. Schnable1,4,5,7 1Bioinformatics and Computational Biology Graduate Program, Iowa State University, Ames, Iowa 50010, USA; 2Department of Electrical and Computer Engineering, Iowa State University, Ames, Iowa 50010, USA; 3Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA; 4Interdepartmental Plant Physiology Graduate Major and Department of Genetics, Development, and Cell Biology, Iowa State University, Ames, Iowa 50010, USA; 5Department of Agronomy and Center for Plant Genomics, Iowa State University, Ames, Iowa 50010, USA 454 DNA sequencing technology achieves significant throughput relative to traditional approaches. More than 261,000 ESTs were generated by 454 Life Sciences from cDNA isolated using laser capture microdissection (LCM) from the developmentally important shoot apical meristem (SAM) of maize (Zea mays L.). This single sequencing run annotated >25,000 maize genomic sequences and also captured ∼400 expressed transcripts for which homologous sequences have not yet been identified in other species. Approximately 70% of the ESTs generated in this study had not been captured during a previous EST project conducted using a cDNA library constructed from hand-dissected apex tissue that is highly enriched for SAMs. In addition, at least 30% of the 454-ESTs do not align to any of the ∼648,000 extant maize ESTs using conservative alignment criteria. These results indicate that the combination of LCM and the deep sequencing possible with 454 technology enriches for SAM transcripts not present in current EST collections. -

EMBL-EBI Powerpoint Presentation
Processing data from high-throughput sequencing experiments Simon Anders Use-cases for HTS · de-novo sequencing and assembly of small genomes · transcriptome analysis (RNA-Seq, sRNA-Seq, ...) • identifying transcripted regions • expression profiling · Resequencing to find genetic polymorphisms: • SNPs, micro-indels • CNVs · ChIP-Seq, nucleosome positions, etc. · DNA methylation studies (after bisulfite treatment) · environmental sampling (metagenomics) · reading bar codes Use cases for HTS: Bioinformatics challenges Established procedures may not be suitable. New algorithms are required for · assembly · alignment · statistical tests (counting statistics) · visualization · segmentation · ... Where does Bioconductor come in? Several steps: · Processing of the images and determining of the read sequencest • typically done by core facility with software from the manufacturer of the sequencing machine · Aligning the reads to a reference genome (or assembling the reads into a new genome) • Done with community-developed stand-alone tools. · Downstream statistical analyis. • Write your own scripts with the help of Bioconductor infrastructure. Solexa standard workflow SolexaPipeline · "Firecrest": Identifying clusters ⇨ typically 15..20 mio good clusters per lane · "Bustard": Base calling ⇨ sequence for each cluster, with Phred-like scores · "Eland": Aligning to reference Firecrest output Large tab-separated text files with one row per identified cluster, specifying · lane index and tile index · x and y coordinates of cluster on tile · for each -

Webnetcoffee
Hu et al. BMC Bioinformatics (2018) 19:422 https://doi.org/10.1186/s12859-018-2443-4 SOFTWARE Open Access WebNetCoffee: a web-based application to identify functionally conserved proteins from Multiple PPI networks Jialu Hu1,2, Yiqun Gao1, Junhao He1, Yan Zheng1 and Xuequn Shang1* Abstract Background: The discovery of functionally conserved proteins is a tough and important task in system biology. Global network alignment provides a systematic framework to search for these proteins from multiple protein-protein interaction (PPI) networks. Although there exist many web servers for network alignment, no one allows to perform global multiple network alignment tasks on users’ test datasets. Results: Here, we developed a web server WebNetcoffee based on the algorithm of NetCoffee to search for a global network alignment from multiple networks. To build a series of online test datasets, we manually collected 218,339 proteins, 4,009,541 interactions and many other associated protein annotations from several public databases. All these datasets and alignment results are available for download, which can support users to perform algorithm comparison and downstream analyses. Conclusion: WebNetCoffee provides a versatile, interactive and user-friendly interface for easily running alignment tasks on both online datasets and users’ test datasets, managing submitted jobs and visualizing the alignment results through a web browser. Additionally, our web server also facilitates graphical visualization of induced subnetworks for a given protein and its neighborhood. To the best of our knowledge, it is the first web server that facilitates the performing of global alignment for multiple PPI networks. Availability: http://www.nwpu-bioinformatics.com/WebNetCoffee Keywords: Multiple network alignment, Webserver, PPI networks, Protein databases, Gene ontology Background tools [7–10] have been developed to understand molec- Proteins are involved in almost all life processes. -

An Open-Sourced Bioinformatic Pipeline for the Processing of Next-Generation Sequencing Derived Nucleotide Reads
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.20.050369; this version posted May 28, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. An open-sourced bioinformatic pipeline for the processing of Next-Generation Sequencing derived nucleotide reads: Identification and authentication of ancient metagenomic DNA Thomas C. Collin1, *, Konstantina Drosou2, 3, Jeremiah Daniel O’Riordan4, Tengiz Meshveliani5, Ron Pinhasi6, and Robin N. M. Feeney1 1School of Medicine, University College Dublin, Ireland 2Division of Cell Matrix Biology Regenerative Medicine, University of Manchester, United Kingdom 3Manchester Institute of Biotechnology, School of Earth and Environmental Sciences, University of Manchester, United Kingdom [email protected] 5Institute of Paleobiology and Paleoanthropology, National Museum of Georgia, Tbilisi, Georgia 6Department of Evolutionary Anthropology, University of Vienna, Austria *Corresponding Author Abstract The emerging field of ancient metagenomics adds to these Bioinformatic pipelines optimised for the processing and as- processing complexities with the need for additional steps sessment of metagenomic ancient DNA (aDNA) are needed in the separation and authentication of ancient sequences from modern sequences. Currently, there are few pipelines for studies that do not make use of high yielding DNA cap- available for the analysis of ancient metagenomic DNA ture techniques. These bioinformatic pipelines are tradition- 1 4 ally optimised for broad aDNA purposes, are contingent on (aDNA) ≠ The limited number of bioinformatic pipelines selection biases and are associated with high costs. -

Sequence Alignment/Map Format Specification
Sequence Alignment/Map Format Specification The SAM/BAM Format Specification Working Group 3 Jun 2021 The master version of this document can be found at https://github.com/samtools/hts-specs. This printing is version 53752fa from that repository, last modified on the date shown above. 1 The SAM Format Specification SAM stands for Sequence Alignment/Map format. It is a TAB-delimited text format consisting of a header section, which is optional, and an alignment section. If present, the header must be prior to the alignments. Header lines start with `@', while alignment lines do not. Each alignment line has 11 mandatory fields for essential alignment information such as mapping position, and variable number of optional fields for flexible or aligner specific information. This specification is for version 1.6 of the SAM and BAM formats. Each SAM and BAMfilemay optionally specify the version being used via the @HD VN tag. For full version history see Appendix B. Unless explicitly specified elsewhere, all fields are encoded using 7-bit US-ASCII 1 in using the POSIX / C locale. Regular expressions listed use the POSIX / IEEE Std 1003.1 extended syntax. 1.1 An example Suppose we have the following alignment with bases in lowercase clipped from the alignment. Read r001/1 and r001/2 constitute a read pair; r003 is a chimeric read; r004 represents a split alignment. Coor 12345678901234 5678901234567890123456789012345 ref AGCATGTTAGATAA**GATAGCTGTGCTAGTAGGCAGTCAGCGCCAT +r001/1 TTAGATAAAGGATA*CTG +r002 aaaAGATAA*GGATA +r003 gcctaAGCTAA +r004 ATAGCT..............TCAGC -r003 ttagctTAGGC -r001/2 CAGCGGCAT The corresponding SAM format is:2 1Charset ANSI X3.4-1968 as defined in RFC1345. -

The Biogrid Interaction Database
D470–D478 Nucleic Acids Research, 2015, Vol. 43, Database issue Published online 26 November 2014 doi: 10.1093/nar/gku1204 The BioGRID interaction database: 2015 update Andrew Chatr-aryamontri1, Bobby-Joe Breitkreutz2, Rose Oughtred3, Lorrie Boucher2, Sven Heinicke3, Daici Chen1, Chris Stark2, Ashton Breitkreutz2, Nadine Kolas2, Lara O’Donnell2, Teresa Reguly2, Julie Nixon4, Lindsay Ramage4, Andrew Winter4, Adnane Sellam5, Christie Chang3, Jodi Hirschman3, Chandra Theesfeld3, Jennifer Rust3, Michael S. Livstone3, Kara Dolinski3 and Mike Tyers1,2,4,* 1Institute for Research in Immunology and Cancer, Universite´ de Montreal,´ Montreal,´ Quebec H3C 3J7, Canada, 2The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada, 3Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544, USA, 4School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JR, UK and 5Centre Hospitalier de l’UniversiteLaval´ (CHUL), Quebec,´ Quebec´ G1V 4G2, Canada Received September 26, 2014; Revised November 4, 2014; Accepted November 5, 2014 ABSTRACT semi-automated text-mining approaches, and to en- hance curation quality control. The Biological General Repository for Interaction Datasets (BioGRID: http://thebiogrid.org) is an open access database that houses genetic and protein in- INTRODUCTION teractions curated from the primary biomedical lit- Massive increases in high-throughput DNA sequencing erature for all major model organism species and technologies (1) have enabled an unprecedented level of humans. As of September 2014, the BioGRID con- genome annotation for many hundreds of species (2–6), tains 749 912 interactions as drawn from 43 149 pub- which has led to tremendous progress in the understand- lications that represent 30 model organisms. -

Multi-Scale Analysis and Clustering of Co-Expression Networks
Multi-scale analysis and clustering of co-expression networks Nuno R. Nen´e∗1 1Department of Genetics, University of Cambridge, Cambridge, UK September 14, 2018 Abstract 1 Introduction The increasing capacity of high-throughput genomic With the advent of technologies allowing the collec- technologies for generating time-course data has tion of time-series in cell biology, the rich structure stimulated a rich debate on the most appropriate of the paths that cells take in expression space be- methods to highlight crucial aspects of data struc- came amenable to processing. Several methodologies ture. In this work, we address the problem of sparse have been crucial to carefully organize the wealth of co-expression network representation of several time- information generated by experiments [1], including course stress responses in Saccharomyces cerevisiae. network-based approaches which constitute an excel- We quantify the information preserved from the origi- lent and flexible option for systems-level understand- nal datasets under a graph-theoretical framework and ing [1,2,3]. The use of graphs for expression anal- evaluate how cross-stress features can be identified. ysis is inherently an attractive proposition for rea- This is performed both from a node and a network sons related to sparsity, which simplifies the cumber- community organization point of view. Cluster anal- some analysis of large datasets, in addition to being ysis, here viewed as a problem of network partition- mathematically convenient (see examples in Fig.1). ing, is achieved under state-of-the-art algorithms re- The properties of co-expression networks might re- lying on the properties of stochastic processes on the veal a myriad of aspects pertaining to the impact of constructed graphs. -

The Uniprot Knowledgebase BLAST
Introduction to bioinformatics The UniProt Knowledgebase BLAST UniProtKB Basic Local Alignment Search Tool A CRITICAL GUIDE 1 Version: 1 August 2018 A Critical Guide to BLAST BLAST Overview This Critical Guide provides an overview of the BLAST similarity search tool, Briefly examining the underlying algorithm and its rise to popularity. Several WeB-based and stand-alone implementations are reviewed, and key features of typical search results are discussed. Teaching Goals & Learning Outcomes This Guide introduces concepts and theories emBodied in the sequence database search tool, BLAST, and examines features of search outputs important for understanding and interpreting BLAST results. On reading this Guide, you will Be aBle to: • search a variety of Web-based sequence databases with different query sequences, and alter search parameters; • explain a range of typical search parameters, and the likely impacts on search outputs of changing them; • analyse the information conveyed in search outputs and infer the significance of reported matches; • examine and investigate the annotations of reported matches, and their provenance; and • compare the outputs of different BLAST implementations and evaluate the implications of any differences. finding short words – k-tuples – common to the sequences Being 1 Introduction compared, and using heuristics to join those closest to each other, including the short mis-matched regions Between them. BLAST4 was the second major example of this type of algorithm, From the advent of the first molecular sequence repositories in and rapidly exceeded the popularity of FastA, owing to its efficiency the 1980s, tools for searching dataBases Became essential. DataBase searching is essentially a ‘pairwise alignment’ proBlem, in which the and Built-in statistics. -

Functionfinderspresnotes.Pdf
1 A series of three bases is called a codon which translates into an amino acid. A series of amino acids make a protein. The codon wheel is central to this activity as it translates the DNA sequence into amino acids. To use the wheel you must work from the inside circle out to the outer circle. For example if the first triplet of the sequence is CAT, the amino acid it codes for is Histidine (H). 2 Stress to the students that the amino acid sequence is not the entire amino acid sequence, but a region that we can use to search online databases. Normally, amino acid sequences can be comprised of hundreds of amino acids. By following the link to the Uniprot website at the bottom of the protein profile students will be able to see the full sequence of the protein (if time). 3 4 Before discussing the results, stress to the students that the amino acid sequence is not the entire amino acid sequence, but a region that we can use to search online dbdatabases. Normally, amino acid sequences can be comprised of hundreds of amino acids. By following the link to the Uniprot website at the bottom of the protein profile students will be able to see the full sequence of the protein (if time). The antifreeze protein is essential to prevent organisms like the wolf fish from freezing in icy conditions. Q. Is this just exclusive to this one species / what other species may use antifreeze proteins? Antifreeze proteins are found in other organisms including invertebrates such as the “Snow flea” or springtail, as well as species of plant and some bacteria. -

NGS Raw Data Analysis
NGS raw data analysis Introduc3on to Next-Generaon Sequencing (NGS)-data Analysis Laura Riva & Maa Pelizzola Outline 1. Descrip7on of sequencing plaorms 2. Alignments, QC, file formats, data processing, and tools of general use 3. Prac7cal lesson Illumina Sequencing Single base extension with incorporaon of fluorescently labeled nucleodes Library preparaon Automated cluster genera3on DNA fragmenta3on and adapter ligaon Aachment to the flow cell Cluster generaon by bridge amplifica3on of DNA fragments Sequencing Illumina Output Quality Scores Sequence Files Comparison of some exisng plaGorms 454 Ti RocheT Illumina HiSeqTM ABI 5500 (SOLiD) 2000 Amplificaon Emulsion PCR Bridge PCR Emulsion PCR Sequencing Pyrosequencing Reversible Ligaon-based reac7on terminators sequencing Paired ends/sep Yes/3kb Yes/200 bp Yes/3 kb Read length 400 bp 100 bp 75 bp Advantages Short run 7mes. The most popular Good base call Longer reads plaorm accuracy. Good improve mapping in mulplexing repe7ve regions. capability Ability to detect large structural variaons Disadvantages High reagent cost. Higher error rates in repeat sequences The Illumina HiSeq! Stacey Gabriel Libraries, lanes, and flowcells Flowcell Lanes Illumina Each reaction produces a unique Each NGS machine processes a library of DNA fragments for single flowcell containing several sequencing. independent lanes during a single sequencing run Applications of Next-Generation Sequencing Basic data analysis concepts: • Raw data analysis= image processing and base calling • Understanding Fastq • Understanding Quality scores • Quality control and read cleaning • Alignment to the reference genome • Understanding SAM/BAM formats • Samtools • Bedtools Understanding FASTQ file format FASTQ format is a text-based format for storing a biological sequence and its corresponding quality scores. -
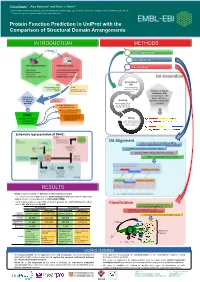
Protein Function Prediction in Uniprot with the Comparison of Structural Domain Arrangements
Tunca Dogan1,*, Alex Bateman1 and Maria J. Martin1 1 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK * To whom correspondence should be addressed: [email protected] Protein Function Prediction in UniProt with the Comparison of Structural Domain Arrangements INTRODUCTION METHODS Data Preparation DA alignment Classification DA Generation Generation of SAAS DAs (Automatic decision tree- UniRule using InterProScan based rule-generating (Manually curated rules results on UniProtKB system) created by curation team) proteins Definition of domain architecture (DA): concatenation of the Requirement of InterPro IDs of the new approaches Annotations from Grouping for automatic other sources domains on the protein annotation proteins under shared sequence Treats domain hits as DAs & separation of strings (instead of a.a.) learning and test sets Pairwise alignment of DAs for similarity detection DAAC Supervised classification (Domain Architecture of query sequences into Mining functional classes Alignment and functional annotations Classification) from source databases (for learning set) Schematic representation of DAAC: DA Alignment A modified version of Needleman-Wunsch global sequence alignment algorithm to compare the DAs by: - treating the domains as the strings in a sequence - working for 7497 InterPro domains instead of 20 a.a - fast due to reduced total number of operations RESULTS • DAAC system is trained for GO term and EC number prediction