Webnetcoffee
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Uniprot at EMBL-EBI's Role in CTTV
Barbara P. Palka, Daniel Gonzalez, Edd Turner, Xavier Watkins, Maria J. Martin, Claire O’Donovan European Bioinformatics Institute (EMBL-EBI), European Molecular Biology Laboratory, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK UniProt at EMBL-EBI’s role in CTTV: contributing to improved disease knowledge Introduction The mission of UniProt is to provide the scientific community with a The Centre for Therapeutic Target Validation (CTTV) comprehensive, high quality and freely accessible resource of launched in Dec 2015 a new web platform for life- protein sequence and functional information. science researchers that helps them identify The UniProt Knowledgebase (UniProtKB) is the central hub for the collection of therapeutic targets for new and repurposed medicines. functional information on proteins, with accurate, consistent and rich CTTV is a public-private initiative to generate evidence on the annotation. As much annotation information as possible is added to each validity of therapeutic targets based on genome-scale experiments UniProtKB record and this includes widely accepted biological ontologies, and analysis. CTTV is working to create an R&D framework that classifications and cross-references, and clear indications of the quality of applies to a wide range of human diseases, and is committed to annotation in the form of evidence attribution of experimental and sharing its data openly with the scientific community. CTTV brings computational data. together expertise from four complementary institutions: GSK, Biogen, EMBL-EBI and Wellcome Trust Sanger Institute. UniProt’s disease expert curation Q5VWK5 (IL23R_HUMAN) This section provides information on the disease(s) associated with genetic variations in a given protein. The information is extracted from the scientific literature and diseases that are also described in the OMIM database are represented with a controlled vocabulary. -

The ELIXIR Core Data Resources: Fundamental Infrastructure for The
Supplementary Data: The ELIXIR Core Data Resources: fundamental infrastructure for the life sciences The “Supporting Material” referred to within this Supplementary Data can be found in the Supporting.Material.CDR.infrastructure file, DOI: 10.5281/zenodo.2625247 (https://zenodo.org/record/2625247). Figure 1. Scale of the Core Data Resources Table S1. Data from which Figure 1 is derived: Year 2013 2014 2015 2016 2017 Data entries 765881651 997794559 1726529931 1853429002 2715599247 Monthly user/IP addresses 1700660 2109586 2413724 2502617 2867265 FTEs 270 292.65 295.65 289.7 311.2 Figure 1 includes data from the following Core Data Resources: ArrayExpress, BRENDA, CATH, ChEBI, ChEMBL, EGA, ENA, Ensembl, Ensembl Genomes, EuropePMC, HPA, IntAct /MINT , InterPro, PDBe, PRIDE, SILVA, STRING, UniProt ● Note that Ensembl’s compute infrastructure physically relocated in 2016, so “Users/IP address” data are not available for that year. In this case, the 2015 numbers were rolled forward to 2016. ● Note that STRING makes only minor releases in 2014 and 2016, in that the interactions are re-computed, but the number of “Data entries” remains unchanged. The major releases that change the number of “Data entries” happened in 2013 and 2015. So, for “Data entries” , the number for 2013 was rolled forward to 2014, and the number for 2015 was rolled forward to 2016. The ELIXIR Core Data Resources: fundamental infrastructure for the life sciences 1 Figure 2: Usage of Core Data Resources in research The following steps were taken: 1. API calls were run on open access full text articles in Europe PMC to identify articles that mention Core Data Resource by name or include specific data record accession numbers. -

The Biogrid Interaction Database
D470–D478 Nucleic Acids Research, 2015, Vol. 43, Database issue Published online 26 November 2014 doi: 10.1093/nar/gku1204 The BioGRID interaction database: 2015 update Andrew Chatr-aryamontri1, Bobby-Joe Breitkreutz2, Rose Oughtred3, Lorrie Boucher2, Sven Heinicke3, Daici Chen1, Chris Stark2, Ashton Breitkreutz2, Nadine Kolas2, Lara O’Donnell2, Teresa Reguly2, Julie Nixon4, Lindsay Ramage4, Andrew Winter4, Adnane Sellam5, Christie Chang3, Jodi Hirschman3, Chandra Theesfeld3, Jennifer Rust3, Michael S. Livstone3, Kara Dolinski3 and Mike Tyers1,2,4,* 1Institute for Research in Immunology and Cancer, Universite´ de Montreal,´ Montreal,´ Quebec H3C 3J7, Canada, 2The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, Ontario M5G 1X5, Canada, 3Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ 08544, USA, 4School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JR, UK and 5Centre Hospitalier de l’UniversiteLaval´ (CHUL), Quebec,´ Quebec´ G1V 4G2, Canada Received September 26, 2014; Revised November 4, 2014; Accepted November 5, 2014 ABSTRACT semi-automated text-mining approaches, and to en- hance curation quality control. The Biological General Repository for Interaction Datasets (BioGRID: http://thebiogrid.org) is an open access database that houses genetic and protein in- INTRODUCTION teractions curated from the primary biomedical lit- Massive increases in high-throughput DNA sequencing erature for all major model organism species and technologies (1) have enabled an unprecedented level of humans. As of September 2014, the BioGRID con- genome annotation for many hundreds of species (2–6), tains 749 912 interactions as drawn from 43 149 pub- which has led to tremendous progress in the understand- lications that represent 30 model organisms. -

PINOT: an Intuitive Resource for Integrating Protein-Protein Interactions James E
Tomkins et al. Cell Communication and Signaling (2020) 18:92 https://doi.org/10.1186/s12964-020-00554-5 METHODOLOGY Open Access PINOT: an intuitive resource for integrating protein-protein interactions James E. Tomkins1, Raffaele Ferrari2, Nikoleta Vavouraki1, John Hardy2,3,4,5,6, Ruth C. Lovering7, Patrick A. Lewis1,2,8, Liam J. McGuffin9* and Claudia Manzoni1,10* Abstract Background: The past decade has seen the rise of omics data for the understanding of biological systems in health and disease. This wealth of information includes protein-protein interaction (PPI) data derived from both low- and high-throughput assays, which are curated into multiple databases that capture the extent of available information from the peer-reviewed literature. Although these curation efforts are extremely useful, reliably downloading and integrating PPI data from the variety of available repositories is challenging and time consuming. Methods: We here present a novel user-friendly web-resource called PINOT (Protein Interaction Network Online Tool; available at http://www.reading.ac.uk/bioinf/PINOT/PINOT_form.html) to optimise the collection and processing of PPI data from IMEx consortium associated repositories (members and observers) and WormBase, for constructing, respectively, human and Caenorhabditis elegans PPI networks. Results: Users submit a query containing a list of proteins of interest for which PINOT extracts data describing PPIs. At every query submission PPI data are downloaded, merged and quality assessed. Then each PPI is confidence scored based on the number of distinct methods used for interaction detection and the number of publications that report the specific interaction. Examples of how PINOT can be applied are provided to highlight the performance, ease of use and potential utility of this tool. -

The Uniprot Knowledgebase BLAST
Introduction to bioinformatics The UniProt Knowledgebase BLAST UniProtKB Basic Local Alignment Search Tool A CRITICAL GUIDE 1 Version: 1 August 2018 A Critical Guide to BLAST BLAST Overview This Critical Guide provides an overview of the BLAST similarity search tool, Briefly examining the underlying algorithm and its rise to popularity. Several WeB-based and stand-alone implementations are reviewed, and key features of typical search results are discussed. Teaching Goals & Learning Outcomes This Guide introduces concepts and theories emBodied in the sequence database search tool, BLAST, and examines features of search outputs important for understanding and interpreting BLAST results. On reading this Guide, you will Be aBle to: • search a variety of Web-based sequence databases with different query sequences, and alter search parameters; • explain a range of typical search parameters, and the likely impacts on search outputs of changing them; • analyse the information conveyed in search outputs and infer the significance of reported matches; • examine and investigate the annotations of reported matches, and their provenance; and • compare the outputs of different BLAST implementations and evaluate the implications of any differences. finding short words – k-tuples – common to the sequences Being 1 Introduction compared, and using heuristics to join those closest to each other, including the short mis-matched regions Between them. BLAST4 was the second major example of this type of algorithm, From the advent of the first molecular sequence repositories in and rapidly exceeded the popularity of FastA, owing to its efficiency the 1980s, tools for searching dataBases Became essential. DataBase searching is essentially a ‘pairwise alignment’ proBlem, in which the and Built-in statistics. -

Biocuration Experts on the Impact of Duplication and Other Data Quality Issues in Biological Databases
Genomics Proteomics Bioinformatics 18 (2020) 91–103 Genomics Proteomics Bioinformatics www.elsevier.com/locate/gpb www.sciencedirect.com PERSPECTIVE Quality Matters: Biocuration Experts on the Impact of Duplication and Other Data Quality Issues in Biological Databases Qingyu Chen 1,*, Ramona Britto 2, Ivan Erill 3, Constance J. Jeffery 4, Arthur Liberzon 5, Michele Magrane 2, Jun-ichi Onami 6,7, Marc Robinson-Rechavi 8,9, Jana Sponarova 10, Justin Zobel 1,*, Karin Verspoor 1,* 1 School of Computing and Information Systems, University of Melbourne, Melbourne, VIC 3010, Australia 2 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Cambridge CB10 1SD, UK 3 Department of Biological Sciences, University of Maryland, Baltimore, MD 21250, USA 4 Department of Biological Sciences, University of Illinois at Chicago, Chicago, IL 60607, USA 5 Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA 6 Japan Science and Technology Agency, National Bioscience Database Center, Tokyo 102-8666, Japan 7 National Institute of Health Sciences, Tokyo 158-8501, Japan 8 Swiss Institute of Bioinformatics, CH-1015 Lausanne, Switzerland 9 Department of Ecology and Evolution, University of Lausanne, CH-1015 Lausanne, Switzerland 10 Nebion AG, 8048 Zurich, Switzerland Received 8 December 2017; revised 24 October 2018; accepted 14 December 2018 Available online 9 July 2020 Handled by Zhang Zhang Introduction assembled, annotated, and ultimately submitted to primary nucleotide databases such as GenBank [2], European Nucleo- tide Archive (ENA) [3], and DNA Data Bank of Japan Biological databases represent an extraordinary collective vol- (DDBJ) [4] (collectively known as the International Nucleotide ume of work. -

Functionfinderspresnotes.Pdf
1 A series of three bases is called a codon which translates into an amino acid. A series of amino acids make a protein. The codon wheel is central to this activity as it translates the DNA sequence into amino acids. To use the wheel you must work from the inside circle out to the outer circle. For example if the first triplet of the sequence is CAT, the amino acid it codes for is Histidine (H). 2 Stress to the students that the amino acid sequence is not the entire amino acid sequence, but a region that we can use to search online databases. Normally, amino acid sequences can be comprised of hundreds of amino acids. By following the link to the Uniprot website at the bottom of the protein profile students will be able to see the full sequence of the protein (if time). 3 4 Before discussing the results, stress to the students that the amino acid sequence is not the entire amino acid sequence, but a region that we can use to search online dbdatabases. Normally, amino acid sequences can be comprised of hundreds of amino acids. By following the link to the Uniprot website at the bottom of the protein profile students will be able to see the full sequence of the protein (if time). The antifreeze protein is essential to prevent organisms like the wolf fish from freezing in icy conditions. Q. Is this just exclusive to this one species / what other species may use antifreeze proteins? Antifreeze proteins are found in other organisms including invertebrates such as the “Snow flea” or springtail, as well as species of plant and some bacteria. -
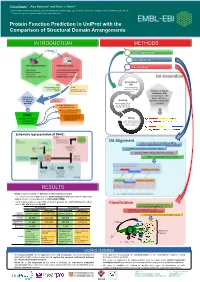
Protein Function Prediction in Uniprot with the Comparison of Structural Domain Arrangements
Tunca Dogan1,*, Alex Bateman1 and Maria J. Martin1 1 European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK * To whom correspondence should be addressed: [email protected] Protein Function Prediction in UniProt with the Comparison of Structural Domain Arrangements INTRODUCTION METHODS Data Preparation DA alignment Classification DA Generation Generation of SAAS DAs (Automatic decision tree- UniRule using InterProScan based rule-generating (Manually curated rules results on UniProtKB system) created by curation team) proteins Definition of domain architecture (DA): concatenation of the Requirement of InterPro IDs of the new approaches Annotations from Grouping for automatic other sources domains on the protein annotation proteins under shared sequence Treats domain hits as DAs & separation of strings (instead of a.a.) learning and test sets Pairwise alignment of DAs for similarity detection DAAC Supervised classification (Domain Architecture of query sequences into Mining functional classes Alignment and functional annotations Classification) from source databases (for learning set) Schematic representation of DAAC: DA Alignment A modified version of Needleman-Wunsch global sequence alignment algorithm to compare the DAs by: - treating the domains as the strings in a sequence - working for 7497 InterPro domains instead of 20 a.a - fast due to reduced total number of operations RESULTS • DAAC system is trained for GO term and EC number prediction -

Uniprot.Ws: R Interface to Uniprot Web Services
Package ‘UniProt.ws’ September 26, 2021 Type Package Title R Interface to UniProt Web Services Version 2.33.0 Depends methods, utils, RSQLite, RCurl, BiocGenerics (>= 0.13.8) Imports AnnotationDbi, BiocFileCache, rappdirs Suggests RUnit, BiocStyle, knitr Description A collection of functions for retrieving, processing and repackaging the UniProt web services. Collate AllGenerics.R AllClasses.R getFunctions.R methods-select.R utilities.R License Artistic License 2.0 biocViews Annotation, Infrastructure, GO, KEGG, BioCarta VignetteBuilder knitr LazyLoad yes git_url https://git.bioconductor.org/packages/UniProt.ws git_branch master git_last_commit 5062003 git_last_commit_date 2021-05-19 Date/Publication 2021-09-26 Author Marc Carlson [aut], Csaba Ortutay [ctb], Bioconductor Package Maintainer [aut, cre] Maintainer Bioconductor Package Maintainer <[email protected]> R topics documented: UniProt.ws-objects . .2 UNIPROTKB . .4 utilities . .8 Index 11 1 2 UniProt.ws-objects UniProt.ws-objects UniProt.ws objects and their related methods and functions Description UniProt.ws is the base class for interacting with the Uniprot web services from Bioconductor. In much the same way as an AnnotationDb object allows acces to select for many other annotation packages, UniProt.ws is meant to allow usage of select methods and other supporting methods to enable the easy extraction of data from the Uniprot web services. select, columns and keys are used together to extract data via an UniProt.ws object. columns shows which kinds of data can be returned for the UniProt.ws object. keytypes allows the user to discover which keytypes can be passed in to select or keys via the keytype argument. keys returns keys for the database contained in the UniProt.ws object . -

Automated Data Integration for Developmental Biological Research
REVIEW 3227 Development 134, 3227-3238 (2007) doi:10.1242/dev.001073 Automated data integration for developmental biological research Weiwei Zhong and Paul W. Sternberg In an era exploding with genome-scale data, a major challenge time and place, look up all papers on transcription factors and for developmental biologists is how to extract significant clues determine if there is any overlap. Of course, these searches are easier from these publicly available data to benefit our studies of if there is a comprehensive database that contains this information. individual genes, and how to use them to improve our The process of combining two or more data sets to identify their understanding of development at a systems level. Several intersection is the simplest form of data integration. studies have successfully demonstrated new approaches to Although data integration does not actually create new classic developmental questions by computationally integrating information, it can create new knowledge for the individual; as various genome-wide data sets. Such computational approaches discussed above, it can limit the number of candidates a researcher have shown great potential for facilitating research: instead of should test, thereby allowing more time for an intensive analysis of testing 20,000 genes, researchers might test 200 to the same each candidate. This strategy becomes even more desirable when a effect. We discuss the nature and state of this art as it applies to developmental process or the gene of interest does not have an easily developmental research. screenable phenotype. More-advanced techniques of data integration employ Introduction sophisticated statistical models to improve the extraction of The advent of high-throughput technologies (see glossary, Box 1) meaningful information. -

Three-Dimensional Structures of Carbohydrates and Where to Find Them
International Journal of Molecular Sciences Review Three-Dimensional Structures of Carbohydrates and Where to Find Them Sofya I. Scherbinina 1,2,* and Philip V. Toukach 1,* 1 N.D. Zelinsky Institute of Organic Chemistry, Russian Academy of Science, Leninsky prospect 47, 119991 Moscow, Russia 2 Higher Chemical College, D. Mendeleev University of Chemical Technology of Russia, Miusskaya Square 9, 125047 Moscow, Russia * Correspondence: [email protected] (S.I.S.); [email protected] (P.V.T.) Received: 26 September 2020; Accepted: 16 October 2020; Published: 18 October 2020 Abstract: Analysis and systematization of accumulated data on carbohydrate structural diversity is a subject of great interest for structural glycobiology. Despite being a challenging task, development of computational methods for efficient treatment and management of spatial (3D) structural features of carbohydrates breaks new ground in modern glycoscience. This review is dedicated to approaches of chemo- and glyco-informatics towards 3D structural data generation, deposition and processing in regard to carbohydrates and their derivatives. Databases, molecular modeling and experimental data validation services, and structure visualization facilities developed for last five years are reviewed. Keywords: carbohydrate; spatial structure; model build; database; web-tool; glycoinformatics; structure validation; PDB glycans; structure visualization; molecular modeling 1. Introduction Knowledge of carbohydrate spatial (3D) structure is crucial for investigation of glycoconjugate biological activity [1,2], vaccine development [3,4], estimation of ligand-receptor interaction energy [5–7] studies of conformational mobility of macromolecules [8], drug design [9], studies of cell wall construction aspects [10], glycosylation processes [11], and many other aspects of carbohydrate chemistry and biology. Therefore, providing information support for carbohydrate 3D structure is vital for the development of modern glycomics and glycoproteomics. -

Robustness of Plant Quantitative Disease Resistance Is Provided by a Decentralized Immune Network
Robustness of plant quantitative disease resistance is provided by a decentralized immune network Florent Delplacea,1, Carine Huard-Chauveaua,1, Ullrich Dubiellaa,b, Mehdi Khafifa, Eva Alvareza, Gautier Langina, Fabrice Rouxa, Rémi Peyrauda,c, and Dominique Robya,2 aLaboratoire des Interactions Plantes-Microorganismes, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement, CNRS, Université de Toulouse, 31326 Castanet-Tolosan, France; bKWS SAAT SE & Co, 37574 Einbeck, Germany; and ciMean, 31520 Toulouse, France Edited by Paul Schulze-Lefert, Max Planck Institute for Plant Breeding Research, Cologne, Germany, and approved June 15, 2020 (received for review January 3, 2020) Quantitative disease resistance (QDR) represents the predominant maize wall-associated kinase, ZmWAK1, conferring QDR to form of resistance in natural populations and crops. Surprisingly, northern corn leaf blight, and for which the molecular function very limited information exists on the biomolecular network of the has been associated with the biosynthesis of secondary metabolites signaling machineries underlying this form of plant immunity. This (9, 10). In some cases, NLR genes can also confer QDR (1, 11). lack of information may result from its complex and quantitative Thus, the molecular functions underlying QDR seem to be more nature. Here, we used an integrative approach including geno- diverse than those responsible for qualitative resistance. However, mics, network reconstruction, and mutational analysis to identify for these few identified genes, the identity of the downstream Arabidopsis and validate molecular networks that control QDR in components of QDR and the corresponding gene networks re- thaliana Xanthomonas cam- in response to the bacterial pathogen main largely unknown.