Metalloproteases Affecting Blood Coagulation, Fibrinolysis and Platelet Aggregation from Snake Venoms: Definition and Nomenclature of Interaction Sites
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

B1–Proteases As Molecular Targets of Drug Development
Abstracts B1–Proteases as Molecular Targets of Drug Development B1-001 lin release from the beta cells. Furthermore, GLP-1 also stimu- DPP-IV structure and inhibitor design lates beta cell growth and insulin biosynthesis, inhibits glucagon H. B. Rasmussen1, S. Branner1, N. Wagtmann3, J. R. Bjelke1 and secretion, reduces free fatty acids and delays gastric emptying. A. B. Kanstrup2 GLP-1 has therefore been suggested as a potentially new treat- 1Protein Engineering, Novo Nordisk A/S, Bagsvaerd, Denmark, ment for type 2 diabetes. However, GLP-1 is very rapidly degra- 2Medicinal Chemistry, Novo Nordisk A/S, Maaloev, Denmark, ded in the bloodstream by the enzyme dipeptidyl peptidase IV 3Discovery Biology, Novo Nordisk A/S, Maaloev, DENMARK. (DPP-IV; EC 3.4.14.5). A very promising approach to harvest E-mail: [email protected] the beneficial effect of GLP-1 in the treatment of diabetes is to inhibit the DPP-IV enzyme, thereby enhancing the levels of The incretin hormones GLP-1 and GIP are released from the gut endogenously intact circulating GLP-1. The three dimensional during meals, and serve as enhancers of glucose stimulated insu- structure of human DPP-IV in complex with various inhibitors 138 Abstracts creates a better understanding of the specificity and selectivity of drug-like transition-state inhibitors but can be utilized for the this enzyme and allows for further exploration and design of new design of non-transition-state inhibitors that compete for sub- therapeutic inhibitors. The majority of the currently known DPP- strate binding. Besides carrying out proteolytic activity, the IV inhibitors consist of an alpha amino acid pyrrolidine core, to ectodomain of memapsin 2 also interacts with APP leading to which substituents have been added to optimize affinity, potency, the endocytosis of both proteins into the endosomes where APP enzyme selectivity, oral bioavailability, and duration of action. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Jararhagin ECD-Containing Disintegrin Domain: Expression in Escherichia Coli and Inhibition of the Platelet–Collagen Interaction
Archives of Biochemistry and Biophysics Vol. 369, No. 2, September 15, pp. 295–301, 1999 Article ID abbi.1999.1372, available online at http://www.idealibrary.com on Jararhagin ECD-Containing Disintegrin Domain: Expression in Escherichia coli and Inhibition of the Platelet–Collagen Interaction Ana M. Moura-da-Silva,*,†,‡,1 Alex Lı´nica,* Maisa S. Della-Casa,* Aura S. Kamiguti,§ Paulo L. Ho,¶ Julian M. Crampton,† and R. David G. Theakston‡ *Laborato´rio de Imunopatologia and ¶Laborato´rio de Biotecnologia Molecular, Instituto Butantan, Av. Vital Brasil, 1500, 05503-900, Sa˜o Paulo, Brazil; §Department of Haematology, University of Liverpool, Prescot Street, Liverpool, L69 3BX, United Kingdom; and †Molecular Genetics Unit and ‡Alistair Reid Venom Research Unit, Liverpool School of Tropical Medicine, Pembroke Place, L3 5QA, Liverpool, United Kingdom Received May 17, 1999, and in revised form July 6, 1999 demonstrating that these antibodies recognize the Jararhagin, a hemorrhagin from Bothrops jararaca parent jararhagin molecule. Treatment of the fusion venom, is a soluble snake venom component compris- protein with enterokinase, followed by further cap- ing metalloproteinase and disintegrin cysteine-rich ture of the enzyme, resulted in a band of 30 kDa, the domains and, therefore, is structurally closely related expected size for jararhagin-C. Further purification of to the membrane-bound A Disintegrin And Metallo- the cleaved disintegrin using FPLC Mono-Q columns proteinase (ADAMs) protein family. Its hemorrhagic resulted in one fraction capable of efficiently inhibit- activity is associated with the effects of both metallo- ing collagen-induced platelet aggregation in a dose- proteinase and disintegrin domains; the metallopro- dependent manner (IC50 of 8.5 mg/ml). -

Characterization of a Novel Metalloproteinase in Duvernoy's Gland of Rhabdophis Tigrinus Tigrinus
The Journal of Toxicological Sciences, 157 Vol.31, No.2, 157-168, 2006 CHARACTERIZATION OF A NOVEL METALLOPROTEINASE IN DUVERNOY’S GLAND OF RHABDOPHIS TIGRINUS TIGRINUS Koji KOMORI1, Motomi KONISHI1, Yuji MARUTA1, Michihisa TORIBA2, Atsushi SAKAI2, Akira MATSUDA3, Takamitsu HORI3, Mitsuko NAKATANI4, Naoto MINAMINO4 and Toshifumi AKIZAWA1 1Department of Analytical Chemistry, Faculty of Pharmaceutical Sciences, Setsunan University, 45-1 Nagaotogecho, Hirakata, Osaka 573-0101, Japan 2The Japan Snake Institute, 3318 Yabuzuka Ota, Gunma 379-2301, Japan 3Department of Biochemistry, Faculty of Pharmaceutical Sciences, Hiroshima International University, 5-1-1 Hirokoshingai, Kure, Hiroshima 737-0112, Japan 4Department of Pharmacology, National Cardiovascular Center Research Institute, 5-7-1 Fujishirodai, Suita, Osaka 565-8565, Japan (Received January 31, 2006; Accepted February 20, 2006) ABSTRACT — During the characterization of hemorrhagic factor in venom of Rhabdophis tigrinus tigri- nus, so-called Yamakagashi in Japan, one of the Colubridae family, a novel metalloproteinase with molec- ular weight of 38 kDa in the Duvernoy’s gland of Yamakagashi was identified by gelatin zymography and by monitoring its proteolytic activity using a fluorescence peptide substrate, MOCAc-PLGLA2pr(Dnp)AR-NH2, which was developed for measuring the well-known matrix metalloproteinase (MMP) activity. After purification by gel filtration HPLC and/or column switch HPLC system consisting of an affin- ity column, which was immobilized with a synthetic BS-10 peptide (MQKPRCGVPD) originating from propeptide domain of MMP-7 and a reversed-phase column, the N-terminal amino acid sequence of the 38 kDa metalloproteinase was identified as FNTFPGDLK which shared a high homology to Xenopus MMP-9. The 38 kDa metalloproteinase required Zn2+ and Ca2+ ions for its proteolytic activity. -

Final Program
In Memoriam: Final Program XXV Congress of the International Society on Thrombosis and Haemostasis and 61st Annual SSC Meeting June 20 – 25, 2015 Toronto, Canada www.isth2015.org 1 Final Program Table of Contents 3 Venue and Contacts 5 Invitation and Welcome Message 12 ISTH 2015 Committees 24 Congress Support 25 Sponsors and Exhibitors 27 ISTH Awards 32 ISTH Society Information 37 Program Overview 41 Program Day by Day 55 SSC and Educational Program 83 Master Classes and Career Mentorship Sessions 87 Nurses Forum 93 Scientific Program, Monday, June 22 94 Oral Communications 1 102 Plenary Lecture 103 State of the Art Lectures 105 Oral Communications 2 112 Abstract Symposia 120 Poster Session 189 Scientific Program, Tuesday, June 23 190 Oral Communications 3 198 Plenary Lecture 198 State of the Art Lectures 200 Oral Communications 4 208 Plenary Lecture 209 Abstract Symposia 216 Poster Session 285 Scientific Program, Wednesday, June 24 286 Oral Communications 5 294 Plenary Lecture 294 State of the Art Lectures 296 Oral Communications 6 304 Abstract Symposia 311 Poster Session 381 Scientific Program, Thursday, June 25 382 Oral Communications 7 390 Plenary Lecture 390 Abstract Symposia 397 Highlights of ISTH 399 Exhibition Floor Plan 402 Exhibitor List 405 Congress Information 406 Venue Plan 407 Congress Information 417 Social Program 418 Toronto & Canada Information 421 Transportation in Toronto 423 Future ISTH Meetings and Congresses 2 427 Authors Index 1 Thank You to Everyone Who Supported the Venue and Contacts 2014 World Thrombosis Day -

Final Program N
XXII Congress The International Society on Thrombosis and Haemostasis B July 11-16 2009 O 55th Annual Meeting S of the Scientific and Standardization Committee of the ISTH T O Final Program N Boston - July 11-16 2009 XXII Congress of the International Society on Thrombosis and Haemostasis 2009 Table ISTH of Contents Venue and Contacts 2 Wednesday 209 Welcome Messages 3 – Plenary Lectures 210 Committees 7 – State of the Art Lectures 210 Congress Awards and Grants 15 – Abstract Symposia Lectures 212 Other Meetings 19 – Oral Communications 219 – Posters 239 ISTH Information 20 Program Overview 21 Thursday 305 SSC Meetings and – Plenary Lectures 306 Educational Sessions 43 – State of the Art Lectures 306 – Abstract Symposia Lectures 309 Scientific Program 89 – Oral Communications 316 Monday 90 – Posters 331 – Plenary Lectures 90 Nursing Program 383 – State of the Art Lectures 90 Special Symposia 389 – Abstract Symposia Lectures 92 Satellite Symposia 401 – Oral Communications 100 – Posters 118 Technical Symposia Sessions 411 Exhibition and Sponsors 415 Tuesday 185 – Plenary Lectures 186 Exhibitor and Sponsor Profiles 423 – State of the Art Lectures 186 Congress Information 445 – Abstract Symposia Lectures 188 Map of BCEC 446 – Oral Communications 196 Hotel and Transportation Information 447 ISTH 2009 Congress Information 452 Boston Information 458 Social Events 463 Excursions 465 Authors’ Index 477 1 Venue & Contacts Venue Boston Convention & Exhibition Center 415 Summer Street - Boston, Massachusetts 02210 - USA Phone: +1 617 954 2800 - Fax: +1 617 954 3326 The BCEC is only about 10 minutes by taxi from Boston Logan International Airport. The 2009 Exhibition is located in Hall A and B of the Exhibit Level of the BCEC, along with posters and catering. -

Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif
toxins Article Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif Erika Camacho 1, Libia Sanz 2, Teresa Escalante 1, Alicia Pérez 2, Fabián Villalta 1, Bruno Lomonte 1, Ana Gisele C. Neves-Ferreira 3, Andrés Feoli 1, Juan J. Calvete 2,4, José María Gutiérrez 1 and Alexandra Rucavado 1,* 1 Instituto Clodomiro Picado, Facultad de Microbiología, Universidad de Costa Rica, San José 11501, Costa Rica; [email protected] (E.C.); [email protected] (T.E.); [email protected] (F.V.); [email protected] (B.L.); [email protected] (A.F.); [email protected] (J.M.G.) 2 Instituto de Biomedicina de Valencia, Consejo Superior de Investigaciones Científicas, Valencia 46010, Spain; [email protected] (L.S.); [email protected] (A.P.); [email protected] (J.J.C.) 3 Laboratório de Toxinologia, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro 21040-900, Brazil; [email protected] 4 Departamento de Biotecnología, Universidad Politécnica de Valencia, Valencia 46022, Spain * Correspondence: [email protected]; Tel.: +506-25117876 Academic Editor: Nicholas R. Casewell Received: 12 September 2016; Accepted: 30 September 2016; Published: 12 October 2016 Abstract: Snake venom metalloproteinases (SVMPs) play key biological roles in prey immobilization and digestion. The majority of these activities depend on the hydrolysis of relevant protein substrates in the tissues. Hereby, we describe several isoforms and a cDNA clone sequence, corresponding to PII SVMP homologues from the venom of the Central American pit viper Bothriechis lateralis, which have modifications in the residues of the canonical sequence of the zinc-binding motif HEXXHXXGXXH. -

Center for Drug Evaluation and Research Application
CENTER FOR DRUG EVALUATION AND RESEARCH APPLICATION NUMBER: 22-512 PHARMACOLOGY REVIEW(S) Tertiary Pharmacology Review By: Paul C. Brown, Ph.D., ODE Associate Director for Pharmacology and Toxicology OND IO NDA: 22-307 Submission date: 4/19/2010 Drug: Pradaxa™ (Dabigatran etexilate mesylate) Sponsor: Boehringer Ingelheim Pharma GmbH & Co. KG Indication: Prevention of stroke and systemic embolism in patients with atrial fibrillation Reviewing Division: Division of Cardiovascular and Renal Products Comments: The pharm/tox reviewer and supervisor found the nonclinical information submitted for dabigatran to be sufficient to support the proposed use. The reviewer proposes pregnancy category C for the labeling. Dabigatran induced some reproductive toxicity in rats manifest as a decreased number of implantations, decreased number of viable fetuses, increase in the resorption rate, increase in post-implantation loss, and an increase in the number of dead offspring when given at doses of 70 mg/kg (about 2.6 to 3 times the MRHD of 300 mg/day on a mg/m2 basis). Dabigatran also induced some fetal structural variations but did not induce fetal malformations in rats or rabbits. Dabigatran was evaluated for carcinogenicity in 2-year rat and mouse studies. The Executive Carcinogenicity Assessment Committee concluded that these studies were adequate and there were no drug-related tumors in either study. Conclusions: I concur with the Division pharm/tox conclusion that the nonclinical data support approval of this NDA. No additional nonclinical studies are recommended at this time. The proposed Established Pharmacologic Class for dabigatran is "direct thrombin inhibitor". This is appropriate because it is consistent with other moieties of this class. -

Antitumoral Activity of Snake Venom Proteins: New Trends in Cancer Therapy
Hindawi Publishing Corporation BioMed Research International Volume 2014, Article ID 203639, 19 pages http://dx.doi.org/10.1155/2014/203639 Review Article Antitumoral Activity of Snake Venom Proteins: New Trends in Cancer Therapy Leonardo A. Calderon,1 Juliana C. Sobrinho,1 Kayena D. Zaqueo,1 Andrea A. de Moura,1 Amy N. Grabner,1 Maurício V. Mazzi,2 Silvana Marcussi,3 Auro Nomizo,4 CarlaF.C.Fernandes,1 Juliana P. Zuliani,1 Bruna M. A. Carvalho,5 Saulo L. da Silva,5 Rodrigo G. Stábeli,1 and Andreimar M. Soares1 1 Centro de Estudos de Biomoleculas´ Aplicadas aSa` ude,´ CEBio, Fundac¸ao˜ Oswaldo Cruz, Fiocruz Rondoniaˆ e Departamento de Medicina, Universidade Federal de Rondonia,ˆ UNIR, Porto Velho, RO, Brazil 2 Fundac¸ao˜ Herm´ınio Ometto, UNIARARAS, Nucleo´ de Cienciasˆ da Saude-NUCISA,´ 13607-339 Araras, SP, Brazil 3 Departamento de Qu´ımica, Universidade Federal de Lavras, UFLA, 37200-000 Lavras, MG, Brazil 4 Departamento de Analises´ Cl´ınicas, Toxicologicas´ e Bromatologicas,´ Faculdade de Cienciasˆ Farmaceuticasˆ de Ribeirao˜ Preto, Universidade de Sao˜ Paulo, USP, Ribeirao˜ Preto, SP, Brazil 5 Departamento de Qu´ımica, Biotecnologia e Engenharia de Bioprocessos, Universidade Federal de Sao˜ Joao˜ del Rei, UFSJ, Campus Alto paraopeba, Ouro Branco, MG, Brazil Correspondence should be addressed to Andreimar M. Soares; [email protected] Received 20 September 2013; Revised 7 December 2013; Accepted 8 December 2013; Published 13 February 2014 Academic Editor: Fernando Albericio Copyright © 2014 Leonardo A. Calderon et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -
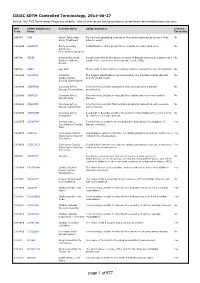
CDISC SDTM Controlled Terminology, 2014-06-27
CDISC SDTM Controlled Terminology, 2014-06-27 Source: NCI EVS Terminology Resources website: http://www.cancer.gov/cancertopics/cancerlibrary/terminologyresources/cdisc NCI CDISC Submission Codelist Name CDISC Definition Codelist Code Value Extensible C66767 ACN Action Taken with Terminology specifying changes to the study treatment as a result of an No Study Treatment adverse event. C101865 ACSPCAT Acute Coronary A classification of the presentation of acute coronary syndrome. No Syndrome Presentation Category C66769 AESEV Severity/Intensity A scale that defines the degree or state of disease existing in a patient as a No Scale for Adverse result of the occurrence of an adverse event. (NCI) Events C66781 AGEU Age Unit Those units of time that are routinely used to express the age of a subject. No C101842 CACRDSC Canadian The anginal classifications as measured by the Canadian Cardiovascular No Cardiovascular Society grading scale. Society Classification C101843 CADPRSN Coronary Artery A terminology codelist associated with coronary artery disease No Disease Presentation presentation. C101849 CADRISK Coronary Artery A terminology codelist to describe the relative risk of coronary artery No Disease Risk disease. C101844 CADSYMP Coronary Artery A terminology codelist that contains symptoms associated with coronary No Disease Symptoms artery disease. C101860 CARTDOM Coronary Artery A codelist to describe whether the posterior descending artery comes from No Dominance the right or left vessel system. C102575 CCINVTYP Contact Case Terminology relevant to the identification and clinical investigation of Yes Investigation Contact disease contacts. Type C101837 CCRCLS Consensus Cardiac Classification system test name for cardiac parameters that are authored or Yes Classification System endorsed by organizations. -

Structural, Functional and Therapeutic Aspects of Snake Venom Metal- Loproteinases
Send Orders for Reprints to [email protected] 28 Mini-Reviews in Organic Chemistry, 2014, 11, 28-44 Structural, Functional and Therapeutic Aspects of Snake Venom Metal- loproteinases P. Chellapandi* Department of Bioinformatics, School of Life Sciences, Bharathidasan University, Tiruchirappalli-620024, Tamil Nadu, India Abstract: Snake venoms are rich sources of metalloproteinases that are of biological interest due to their diverse molecu- lar diversity and selective therapeutic applications. Snake venoms metalloproteinases (SVMPs) belong to the MEROPS peptidase family M12B or reprolysin subfamily, which are consisted of four major domains include a reprolysin catalytic domain, a disintegrin domain, a reprolysin family propeptide domain and a cysteine-rich domain. The appropriate struc- tural and massive sequences information have been available for SVMPs family of enzymes in the Protein Data Bank and National Center for Biotechnology Information, respectively. Functional essentiality of every domain and a crucial contri- bution of binding geometry, primary specificity site, and structural motifs have been studied in details, pointing the way for designing potential anti-coagulation, antitumor, anti-complementary and anti-inflammatory drugs or peptides. These enzymes have been reported to degrade fibrinogen, fibrin and collagens, and to prevent progression of clot formation. An- giotensin-converting enzyme activity, antibacterial properties, haemorrhagic activity and platelet aggregation response of SVMPs have been studied earlier. Structural information of these enzymes together with recombinant DNA technology would strongly promote the construction of many recombinant therapeutic peptides, particularly fibrinogenases and vac- cines. We have comprehensively reviewed the structure-function-evolution relationships of SVMPs family proteins and their advances in the promising target models for structure-based inhibitors and peptides design.