FRAC Code List ©*2020: Fungal Control Agents Sorted by Cross Resistance Pattern and Mode of Action (Including FRAC Code Numbering)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Clinical Epidemiology and Treatment Outcome of Hexaconazole Poisoning – a Prospective Six Year Study
Hexaconazole - Clinical epidemiology and treatment outcome A. Anand & P. Abinash ORIGINAL ARTICLE ORIGINAL ARTICLE Clinical epidemiology and treatment outcome of Hexaconazole poisoning – A prospective six year study ACHARYA ANAND1 and PANDA ABINASH2 1Department of Pharmacology, Konaseema Institute of Medical Sciences & Research Foundation, Amalapuram, Andhra Pradesh, India 2Department of Pharmacology, M.K.C.G. Medical College, Berhampur, Odisha, India. Abstract Background: Hexaconazole is a category 3/4 of poison as per the W.H.O Expert Group on Pesticide Residues. Hexaconazole is used to control infection by fungi in paddy and other crops. Apart from destroying the target species, it can also cause damage to humans. There have been discrete reports of instances of human poisoning due to hexaconazole. Methodology: A patient record-based cross-sectional study was carried out in Konaseema Institute of Medical Science & Research Foundation, Amalapuram, Andhra Pradesh, India during a period from March 2014 to April 2020 on 26 confirmed cases of hexaconazole poisoning. The clinic-demographic data, hematological, and biochemical parameters at the time of admission and at 72 hrs as well as the outcome were recorded and analyzed using descriptive statistics and paired t test. Result: The prevalence of hexaconazole poisoning was 4.79% of all poisoning cases. The major clinical presentation was gastrointestinal symptoms with vomiting being commonest. There was no significant change in the biochemical and hematological parameters. The mean duration of hospitalization was 4.93+1.39 days. The recovery rate was 100% without any major sequel. Conclusion: Poisoning due to hexaconazole is uncommon in comparison to poisoning by other pesticides in the agricultural community. -
Frac List of Fungicide Common Names - 2016
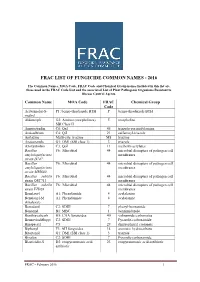
FRAC LIST OF FUNGICIDE COMMON NAMES - 2016 The Common Names, MOA Code, FRAC Code and Chemical Group names included in this list are those used in the FRAC Code List and the associated List of Plant Pathogenic Organisms Resistant to Disease Control Agents. Common Name MOA Code FRAC Chemical Group Code Acibenzolar-S- P1: benzo-thiadiazole BTH P benzo-thiadiazole BTH methyl Aldimorph G2: Amines (morpholines) 5 morpholine SBI Class II Ametoctradin C8: QxI 45 triazolo-pyrimidylamine Amisulbrom C4: QiI 21 sulfamoyl-triazole Anilazine Multi-site: triazine M8 triazine Azaconazole G1: DMI (SBI class 1) 3 triazole Azoxystrobin C3: QoI 11 methoxy-acrylates Bacillus F6: Microbial 44 microbial disrupters of pathogen cell amyloliquefaciens membranes strain D747 Bacillus F6: Microbial 44 microbial disrupters of pathogen cell amyloliquefaciens membranes strain MBI600 Bacillus subtilis F6: Microbial 44 microbial disrupters of pathogen cell strain QST713 membranes Bacillus subtilis F6: Microbial 44 microbial disrupters of pathogen cell strain FZB24 membranes Benalaxyl A1: Phenylamide 4 acylalanine Benalaxyl-M A1: Phenylamide 4 acylalanine (kiralaxyl) Benodanil C2: SDHI 7 phenyl-benzamide Benomyl B1: MBC 1 benzimidazole Benthiavalicarb H5: CAA fungicides 40 valinamide carbamates Benzovindiflupyr C2: SDHI 7 Pyrazole-carboxamide Binapacryl C5 29 dinitrophenyl crotonate Biphenyl F3: AH fungicides 14 aromatic hydrocarbons Bitertanol G1: DMI (SBI class 1) 3 triazole Bixafen C2: SDHI 7 Pyrazole-carboxamide Blasticidin-S D2: enopyranuronic acid 23 enopyranuronic -

Antifungal Drugs Are Those Drugs Which Inhibit Or Retard Fungal Growth
Prepared by- Dr Arpita Shrivastav DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Antifungal drugs are those drugs which inhibit or retard fungal growth. DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Sites of action of antifungal drugs Chitin + Glucan DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA SITE OF ACTION DRUGS Cell membrane Ergosterol binding Amphotericin B, Natamycin and Nystatin Ergosterole synthesis inhibitors Allylamines (Amorolfine, (Squalene epoxidase inhibitor) Terbinafine etc.) Ergosterol synthesis inhibitors Azoles (Ketoconazole, Miconazole (lanosterol to ergosterol) etc.) Cell wall β glucan synthase inhibitor Echinocandins Intracelluler Pyrimidine analogue Flucytosine Mitotic inhibitors Gresiofulvin Other site Benzoic acid , salicylic acid, tolnaftate, castor oil etc. DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Source – Streptomyces nodosus Mechanism of action – Increases permeability of cell membrane by binding to ergosterol Antimicrobial spectrum – broad spectrum Resistance – when less ergosterol in membrane Pharmacokinetics – poorly absorbed from GIT, do not cross BBB Adverse effects - Nephrotoxicity Drug interaction – 1. with miconazole antagonistic effect 2. with flucytosine synergistic effect Dose – Dog -0.25 to 0.50 mg/kg, 3 times weekly Cat – 0.1 to 0.50 mg/kg, 3 times weekly DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Source – Streptomyces noursie Streptomyces aureus Mechanism of action – It binds to ergosteol and forms pores in cell membrane DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Source - Streptomyces natalensis Mechanism of action – Binds to ergosterol and cause leakiness of cell membrane DR ARPITA SHRIVASTAV ASSTT. PROFESSOR VETERINARY COLLEGE REWA Pentameric ring compounds containing at least one other non-carbon atom of either nitrogen, sulfur, or oxygen. -

Group of Fungicides
DEPARTMENT OF PLANT PATHOLOGY UNDER GRADUATE EDUCATION PAT 201 PRINCIPLES OF CROP DISEASE MANAGEMENT (1+1) COURSE TEACHER Dr. V. PRAKASAM PROFOESSOR (PLANT PATHOLOGY) SYLLABUS THEORY Principles of Crop disease management –– Epidemiology of crop diseases – Disease surveillance, assessment of disease intensity - forecasting. Survival and mode of spread of plant pathogens – Types of resistance – Cross protection – mechanism of resistance,. Methods and management of plant diseases - Fungicides – Characteristics of an ideal fungicide - Classification – groups of fungicides – antibiotics. Formulations – compatibility. Phytotoxicity – precautions and safety measures in handling. Management of diseases – seed, soil foliar and post harvest diseases – seed health testing methods – Simple diagnostic techniques for identification of diseases. Biological control and their scope – biocontrol agents – Fungi, bacteria, vesicular arbuscular mycorrhizae – Plant products and antiviral principles. Biotechnological approaches in plant disease management. PRACTICAL Various groups of fungicides and antibiotics – Preparation of Bordeaux mixture and Bordeaux paste – Preparation of fungicidal spray solution – methods of application - seed treatment – Soil and foliar application –– demonstration of Phytotoxicity – Production of immunized seedlings in citrus – Biological control agents- Trichoderma, Pseudomonas and vesicular arbuscular mycorrhizae - Methods of mass production and application. Preparation of botanicals – leaf extracts, oil emulsions and anti viral -

(12) United States Patent (10) Patent No.: US 8,110,689 B2 Wada Et Al
US008110689B2 (12) United States Patent (10) Patent No.: US 8,110,689 B2 Wada et al. (45) Date of Patent: *Feb. 7, 2012 (54) BENZANILIDES WITH INSECTICIDAL M. Waksmundzka-Hajnos, “Chromatographic Separation of Nitro ACTIVITY Phenones and Their Reduced Derivatives on Thin Layers of Polar Adsorbents”, XP-008069069, pp. 159-171. Jun-Ichi Inoh et al., “Palladium-Catalyzed Coupling Reaction of (75) Inventors: Katsuaki Wada, Tochigi (JP); Tetsuya 4-Alkylnitrobenzenes with Aryl Bromides at Their Benzylic Posi Murata, Tochigi (JP); Katsuhiko tion”, XP-004130892, Tetrahedron Letters, 1998, pp. 4673-4676. Shibuya, Tochigi (JP); Eiichi Shimojo, M. Makosza et al., “Synthesis of (p-Nitroaryl)diarylmethanes via Vicarious Nucleophilic Substitution of Hydrogen', XP-002399354. Tochigi (JP) Synthesis, No. 9, 2000, pp. 1237-1240. S. Florio et al., “Vicarious Nucleophilic Substitution of (73) Assignee: Bayer Cropsciene AG, Monheim (DE) (Chloroalkyl)heterocycles with Nitroarenes', XP-002399353, Eur, J. Org. Chem., 2004, pp. 2118-2124. (*) Notice: Subject to any disclaimer, the term of this W. Waiers, “Some Substitution Reactions of patent is extended or adjusted under 35 4-Aminodiphenylmethane', XP-008069039, pp. 1060-1064. G. Esselen, Jr., “The Splitting of Benzhydrols by the Action of Bro U.S.C. 154(b) by 403 days. mine”, XP-002399352, pp. 308-324. This patent is Subject to a terminal dis M.Z.A. Badr et al., “Molecular Rearrangements. 14. Photolysis and claimer. Thermolysis of Phenylpropionanilides”, XP-0023993.51, J. Org. Chem., vol. 44, No. 18, 1979, pp. 3244-3247. Y. Watanabe et al., “Stilbene Derivative. Its Preparation, and (21) Appl. No.: 11/917,411 Electrophotographic Photoreceptor Using Same', XP-002399363, 2000, 2 pgs. -

Modern Fungicides and Antifungal Compounds VI
buchcover_fungizides18-8-2011 18.08.2011 9:51 Uhr Seite 1 C M Y CM MY CY CMY K Persistent Identifier: urn:nbn:de:0294-sp-2011-Reinh-8 Spectrum Phytomedizin Spectrum Phytomedizin Proceedings of the 16th International Reinhardsbrunn Symposium on Modern Fungicides and Antifungal Com- pounds, 2010 The Proceedings of the 16th International Reinhardsbrunn Symposium on Modern Fungicides and Antifungal Com- H.W. Dehne, H.B.Gisi, Deising, U. K.Lyr (Eds.) Kuck, P.E. Russell, H. H. H.W. Dehne, H.B. Deising, U. Gisi, K. H. Kuck, P.E. Russell, H. Lyr (Eds.) pounds uphold the tradition of all previous Symposium Proceedings in this series by bringing you research reports on the increasingly complex discipline of fungicide science. As technology develops and new priorities are set for the Modern Fungicides and control of plant diseases, the research undertaken to provide the tools for disease control expands. In order to Antifungal Compounds VI use these tools efficiently, information must be available on the biological and chemical properties of the disease control agents. The present Proceedings provide a unique insight into current research and are an invaluable source of reference for students and established scientists. Modern Fungicides and Antifungal Compounds VI 16th International Reinhardsbrunn Symposium Spectrum Phytomedizin April 25 – 29, 2010 Friedrichroda, Germany ISBN: 978-3-941261-10-5 Probedruck Persistent Identifier: urn:nbn:de:0294-sp-2011-Reinh-8 DPG Spectrum Phytomedizin H. W. DEHNE, H. B. DEISING, U. GISI, K. H. KUCK, P. E. RUSSELL, H. LYR (EDS) Modern Fungicides and Antifungal Compounds VI Proceedings of the 16th International Reinhardsbrunn Symposium April 25 -29, 2010, Friedrichroda, Germany Persistent Identifier: urn:nbn:de:0294-sp-2011-Reinh-8 ISBN: 978-3-941261-10-5 Das Werk einschließlich aller Teile ist urheberrechtlich geschützt. -

Emerging Contaminants in Groundwater
Emerging contaminants in groundwater Groundwater Science Programme Open Report OR/11/013 O = H2–CH2–P–OH _ OH BRITISH GEOLOGICAL SURVEY GROUNDWATER SCIENCE PROGRAMME OPEN REPORT OR/11/013 Emerging contaminants in groundwater The National Grid and other Ordnance Survey data are used with the permission of the Controller of Her Majesty’s M E Stuart, K Manamsa, J C Talbot and E J Crane Stationery Office. Licence No:100017897/2011. Keywords Report; groundwater; organic micropollutants; pesticides, pesticide metabolites, pharmaceuticals; veterinary medecines; risk assessment. Front cover Structures of selected polar organic micropollutants: bisphenol A, clopyralid, carbamazepine, estradiol, glyphosate metabolite AMPA, metaldehyde, sulfamethoxazole, Bibliographical reference STUART ME, MANAMSA K, TALBOT J C AND CRANE E J. 2011. Emerging contaminants in groundwater. British Geological Survey Open Report, OR/11/013. 123pp. Copyright in materials derived from the British Geological Survey’s work is owned by the Natural Environment Research Council (NERC) and/or the authority that commissioned the work. You may not copy or adapt this publication without first obtaining permission. Contact the BGS Intellectual Property Rights Section, British Geological Survey, Keyworth, e-mail [email protected]. You may quote extracts of a reasonable length without prior permission, provided a full acknowledgement is given of the source of the extract. Maps and diagrams in this book use topography based on Ordnance Survey mapping. © NERC 2011. All rights reserved Keyworth, Nottingham British Geological Survey 2011 BRITISH GEOLOGICAL SURVEY The full range of our publications is available from BGS shops at British Geological Survey offices Nottingham, Edinburgh, London and Cardiff (Welsh publications only) see contact details below or shop online at www.geologyshop.com BGS Central Enquiries Desk 0115 936 3143 Fax 0115 936 3276 The London Information Office also maintains a reference email [email protected] collection of BGS publications including maps for consultation. -
Antifungals of the Azole Type
RISK PROFILE Antifungals of the azole type This concerns azole type molecules used to eradicate or control fungi. They are used, or may potentially be used, for cosmetic purposes as defined in the European Commission database for cosmetic ingredients currently called CosIng. Date of reporting 24.11.201 4 This risk profile deviates in its format from other risk profiles in the series of pharmacological active substances as presented on the webpages of the Norwegian Food Safety Authority (NFSA). This is because the health risk arising because of use in cosmetics of these molecules mainly has to do with a potential worsening of the serious global problem of antimicrobial/antibiotic resistance/cross-resistance. The other risk profiles in this series concerns the toxicity of different cosmetics ingredients. The NSFA base this risk profile mainly on concerns expressed by the European Medicinal Agency (EMA) that on two occasions – in 2005 and 2011 - addressed the unfortunate use of azole type antifungals in cosmetics products. Also the NSFA observes that the same concern has been expressed by the Council of Europe in a publication as from 2008 concerning the use of Active ingredients in cosmetics. Over the years the Norwegian Medicinal Products Agency has strongly encouraged the NFSA forcefully to oppose any use of molecule in cosmetics that might potentially weaken the medicinal anti-fungus armamentarium against life threatening systemic fungal infections in people having a seriously compromised immune system. Over the years, also the European Commission has addressed this issue asking its scientific committee working in the field of cosmetic products - currently called the Scientific Committee on Consumer Safety (SCCS) - for advice. -

National Profile on Chemicals Management in Cambodia
National Background Information MoE GEF UNEP UNITAR NATIONAL PROFILE ON CHEMICALS MANAGEMENT IN CAMBODIA Prepared by: Enabling Activities for Development of a National Plan for Implementation of the Stockholm Convention Funded by: Global Environment Facility Technical Consulted by: United Nation Institute for Training and Research Ministry of Environment December 2004 - 1 - National Background Information TABLE OF CONTENTS TABLE OF CONTENTS ............................................................................................................................2 LIST OF TABLES.......................................................................................................................................4 ABBREVIATION AND ACRONYM .......................................................................................................vii EXECUTIVE SUMMARY........................................................................................................................iix INTRODUCTION.......................................................................................................................................x CHAPTER 1: NATIONAL BACKGROUND INFORMATION..............................................................1 1.1 Physical and Demographic Context......................................................................................................1 1.2 Political/Geographic Structure of the Country ......................................................................................7 1.3 Agricutural and Industrial Sectors -

Use of High Throughput Assays and Computational Tools; Endocrine Disruptor Screening Program; Notice of Availability and Opportunity for Comment’’
NRDC EPA-HQ-OPPT-2015-0305 August, 2015 Comments from the Natural Resources Defense Council On The Document Titled, ‘‘Use of High Throughput Assays and Computational Tools; Endocrine Disruptor Screening Program; Notice of Availability and Opportunity for Comment’’ To the U.S. Environmental Protection Agency Docket No. EPA-HQ-OPPT-2015-0305 August 18, 2015 Background The Natural Resources Defense Council ("NRDC") is a national, non-profit environmental organization of lawyers, scientists, and other professionals. NRDC presents these comments on behalf of our 1.4 million members and online activists. NRDC does not have any financial interest in the topic of these comments. The endocrine system utilizes highly complex, tightly controlled molecular processes for its optimal functioning in the body. The proper balance of hormones can be synchronized in a variety of ways (including direct protein binding, epigenetic alterations, gene activation and silencing), and is essential across the entirety of the life course. Small changes in the perfectly orchestrated symphony of hormone levels can severely disrupt the harmony necessary for critical windows of development (e.g., fetal development, infanthood, childhood, and adolescence), leaving the body vulnerable to a host of negative health outcomes (such as diabetes, cancer, obesity, and reproductive dysfunction). The last decade has seen an exponential increase in the development of computational, biological, and chemical tools capable of increasing both the number of chemicals analyzed and the pace of chemical toxicity evaluation. These tools, including the EPA Toxicity Forecaster (ToxCast™) and the National Institute of Environmental Health Sciences (NIEHS) Tox21 platforms, have the potential to rapidly generate molecular and cellular data for thousands of chemicals at once, and provide an additional stream of useful information that can aide in regulatory decision-making. -

WO 2012/117051 Al 7 September 2012 (07.09.2012) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2012/117051 Al 7 September 2012 (07.09.2012) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A01N3/00 (2006.01) A01N 55/00 (2006.01) kind of national protection available): AE, AG, AL, AM, A01N 43/90 (2006.01) A01P 3/00 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, A01N 63/00 (2006.01) A01P 1/00 (2006.01) CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, A0 43/653 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, (21) International Application Number: KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, PCT/EP2012/053509 MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, (22) International Filing Date: OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, SD, 1 March 2012 (01 .03.2012) SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GH, (30) Priority Data: GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, 11156773. -

FRAC Code List ©*2021: Fungal Control Agents Sorted by Cross Resistance Pattern and Mode of Action (Including Coding for FRAC Groups on Product Labels)
FRAC Code List ©*2021: Fungal control agents sorted by cross resistance pattern and mode of action (including coding for FRAC Groups on product labels) Disclaimer The technical information contained in this publication is provided to CropLife International/RAC members, non-members, the scientific community, and a broader public audience. While CropLife International and the RACs make every effort to present accurate and reliable information in the guidelines, CropLife International and the RACs do not guarantee the accuracy, completeness, efficacy, timeliness, or correct sequencing of such information. CropLife International and the RACs assume no responsibility for consequences resulting from the use of their information, or in any respect for the content of such information, including but not limited to errors or omissions, the accuracy or reasonableness of factual or scientific assumptions, studies or conclusions. Inclusion of active ingredients and products on the RAC Code Lists is based on scientific evaluation of their modes of action; it does not provide any kind of testimonial for the use of a product or a judgment on efficacy. CropLife International and the RACs are not responsible for, and expressly disclaim all liability for, damages of any kind arising out of use, reference to, or reliance on information provided in the guidelines. Listing of chemical classes or modes of action in any of the CropLife International/RAC recommendations must not be interpreted as approval for use of a compound in a given country. Prior to implementation, each user must determine the current registration status in the country of use and strictly adhere to the uses and instructions approved in that country.