Short-Term Forecasts to Inform the Response to the Covid-19 Epidemic in the UK
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Applied AI and Machine Learning in Translational Medicine
This is a repository copy of Looking beyond the hype : applied AI and machine learning in translational medicine. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/151087/ Version: Published Version Article: Toh, T.S., Dondelinger, F. and Wang, D. (2019) Looking beyond the hype : applied AI and machine learning in translational medicine. EBioMedicine. ISSN 2352-3964 https://doi.org/10.1016/j.ebiom.2019.08.027 Reuse This article is distributed under the terms of the Creative Commons Attribution-NonCommercial-NoDerivs (CC BY-NC-ND) licence. This licence only allows you to download this work and share it with others as long as you credit the authors, but you can’t change the article in any way or use it commercially. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ EBIOM-02366; No of Pages 9 EBioMedicine xxx (2019) xxx Contents lists available at ScienceDirect EBioMedicine journal homepage: www.ebiomedicine.com Review Looking beyond the hype: Applied AI and machine learning in translational medicine Tzen S. Toh a, Frank Dondelinger b, Dennis Wang c,d,⁎ a The Medical School, University of Sheffield, Sheffield, UK b Lancaster Medical School, Furness College, Lancaster University, Bailrigg, Lancaster, UK c NIHR Sheffield Biomedical Research Centre, University of Sheffield, Sheffield, UK d Department of Computer Science, University of Sheffield, Sheffield, UK article info abstract Article history: Big data problems are becoming more prevalent for laboratory scientists who look to make clinical impact. -

NHS England Letter Template
OFFICIAL National Medical Directorate NHS England and NHS Improvement Ms Emma Brown Skipton House Area Coroner for Birmingham and 80 London Road Solihull LONDON The Coroner's Court SE1 6LH 50 Newton Street Birmingham B4 6NE Telephone: Email: 20th September 2019 Email: Dear Ms Brown, Re: Regulation 28 Report to Prevent Future Deaths – David Jonothan Jukes (09/10/2018) Thank you for your Regulation 28 Report dated 12 July 2019 concerning the death of David Jonothan Jukes on 09 October 2018. I was very saddened to read of the circumstances surrounding Mr Jukes’s death and would like to extend my condolences to his family and loved ones during this very difficult time. Please note this response will address the matters of concern from a national level and we would expect the local NHS agencies, which I note are copied in to the letter, to address the local concerns raised. Sufficient information regarding arrests was not provided to liaison and diversion practitioner: NHS England and NHS Improvement will work with police forces to ensure that all relevant information from a police perspective is given to the liaison and diversion practitioner, including the nature of the detained person’s presentation at the point of arrest, if relevant. We will commit to a national event by the end of March 2020 to debrief on this matter and any other information sharing/exchange issues that have been raised in other reports. We will invite liaison and diversion practitioners as well as NHS Commissioners and police representatives in order to share lessons -

Abstracts from the Research Symposium for Medical Students and Foundation Doctors 20 November 2019
Abstracts from the Research Symposium for Medical Students and Foundation Doctors 20 November 2019 November 2019 This abstract book has been produced using author-supplied copy and no editing has been undertaken. No responsibility is assumed for any claims, instructions, methods or drug dosages included in the abstracts: it is recommended that these are verified independently. Undergraduate Prizes Best undergraduate oral presentation Socioeconomic differences in cardiovascular disease risk factor prevalence in people with type 2 diabetes in Scotland: a cross-sectional study Edward Whittaker (Supervisor: Prof. Sarah Wild) University of Edinburgh INTRODUCTION: Health inequalities exist in outcomes of diabetes in different socioeconomic groups and these are particularly marked for cardiovascular disease and its risk factors1. The aim of this study was to describe the association between socioeconomic status and prevalence of cardiovascular risk factors (smoking, body mass index, HbA1C, blood pressure and cholesterol) in people with type 2 diabetes in contemporary Scottish data. PATIENTS AND METHODS: A cross-sectional study was performed of 264,011 people with type 2 diabetes in Scotland who were alive on 30/06/16, identified from the population-based diabetes register. Socioeconomic status was defined using quintiles of the area-based SIMD with Q1 and Q5 used to identify the most and least deprived fifths of the population respectively. Logistic regression models adjusted for age, sex, health board, history of cardiovascular disease and duration of diabetes were used to estimate odds ratios (OR) (and 95% confidence intervals) for Q1 compared to Q5 for each risk factor. RESULTS: The mean (SD) age of the study population was 66.7 (12.8) years, 56.1% were men, 23.6% were in Q1 and 15.1% in Q5. -

Embedding Physical Activity in the Undergraduate Curriculum
July 2018 Embedding physical activity in the undergraduate curriculum Commissioned by Public Health England (PHE) and Sport England to embed physical activity in the undergraduate curricula in a sample of medical schools and schools of health in England during 2017 and 2018. This is part of the PHE & Sport England’s Moving Healthcare Professionals Programme Authors on behalf of Exercise Works Ltd Ann B. Gates MRPharmS BPharm (Hons) Ian K. Ritchie FRCSEd Executive Summary July 2018 movement for movement Forewords A final year medical student’s view on the lack of exercise medicine in undergraduate curricula As a medical student about to graduate, I have been left disappointed by the lack of exercise medicine I have been exposed to during my time at medical school. Despite it being a well-established way to prevent, treat and manage illness, exercise medicine does not feature significantly in medical curricula throughout the United Kingdom. As a result, there is lack of knowledge and awareness in the medical community. This issue is not limited to doctors: all allied health care professionals are in a position to influence positive lifestyle changes and all should be exposed to exercise medicine in their undergraduate curricula. We are currently missing out on a fantastic opportunity to empower the health care professionals of the future to be confident in advising patients about utilising exercise to improve their health and quality of life. This commission report demonstrates that physical activity can be successfully integrated into medical school curricula, and that students can encourage and support this being implemented. Katie Marino Final year medical student, University of Sheffield. -
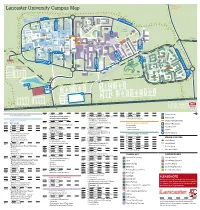
Campus Map CAMPUS
Forrest Hills SOUTH EAST Lancaster University Campus Map CAMPUS NORTH CAMPUS FURNESS AVE B TOWER AVE E C PHYSICS AVE ISO JOHN CREED AVE COUNTY AVE Bailrigg Service Station LANCASTER SQUARE AVE CTP Maintenance GEORGE FOX AVE UNDERPASS Workshops COM PHS WWB County College FYLDE AVE SOUTH CHE CAMPUS D ISS COS The PSC Orchard FAR Bonington Square Step Lancaster TRH Square FAS SBH GFX INF Physics Garden Cycle Route to NORTH DRIVE Fylde College Ellel & Galgate Great Edward SOUTH DRIVE Hall BLN BLM Roberts Court GHC Court Bowland Bowland FUR Wetland North Quad Fylde Grizedale College Quad WEL Furness College Quad Furness Alexandra College Court FYL SAT LIC Square Pendle College Welcome LEC Great Hall Centre CHC Square Reception Engineering F Square Cycle Route to PENDLE AVE ASH Bowland College City Centre BLA Students’ Union ROSSENDALE AVE LIB ENG LSE BLH A Arrival UNH Point University GRIZEDALE AVE House MAN Reception BOWLAND AVE G Graduate College HRB UNDERPASS CPC BOWLAND AVE FARRER AVE GILLOW AVE F Graduate BRH LIBRARY AVE SEC Square A GRADUATE AVE LCC CARTMEL AVE Netball Courts South West I Campus ALEXANDRA PARK DRIVE Barker NORTH WEST RUS House BHF Entrance Lancaster Court House Hotel CAMPUS H Cartmel College Rugby League Pitch PARK BOULEVARD Lacrosse Pitch ECO BARKER HOUSE AVE MED J PRE Lonsdale SOUTH WEST CAMPUS Quad LONSDALE AVE HAZELRIGG LANE Lonsdale College BFB Lake Carter Grass Playing Pitch Astro Turf Pitch L Grass Playing Pitch L Grass Playing Pitch Grass Playing Pitch Grass Playing Pitch 3rd Generation Artificial Pitch Astro Turf -

LU Campus Map.Indd
Lancaster Campus E NORTH CAMPUS SOUTH EAST CAMPUS Forrest Hills 0.5 Miles B C ISO Maintenance E Workshops N S Garage & Fuel Station CTP COM PHS WWB County W College CHE ISS The PS D COS FAR Bonington C Lancaster Step Square TRH Physics FAS SBH GFX INF Garden SOUTH CAMPUS Cycle Route to North Spine Fylde Ellel & Galgate Great Edward College Hall BLN BLM Roberts Court Bowland Court Furness Fylde Grizedale North FUR GHC Bowland College Quad College Quad WEL Quad BLE Pendle Alexandra South Spine Furness FYL SAT College LIC Square College Court Great Welcome CHC Hall Centre Engineering Cycle Square Reception Bowland Square F Route to ASH College City BLA Students’ Union Centre LIB ENG Arrival LEC BLH A Point UNH LSE University House Reception Graduate College E HRB MAN V CPC A E T BRH A U D Graduate A SEC R E G Square G I AV ALEXANDRA EL South A RTM LCC CA West S Campus Entrance RU Lancaster BHF Lacrosse Pitch House Hotel PARK DRI Barker NORTH WEST RD House Cartmel H ULEVA RK BO Court College RA PA ALEXAND H CAMPUS KER O ECO VE Lonsdale BAR U S D Quad E J ME A PRE VE LO NSD Lonsdale ALE Rugby League College A Pitch BFB V Lake Carter Grass Pitch E M Astro Turf L HIO L Grass Pitches Bowling Green SOUTH WEST 3G Artificial Pitch Astro Turf SPC CAMPUS Grass Pitches Cycle K Route to City Grass Playing Pitch Centre Grass Playing Pitch Tennis Courts M6 Junction 33 e ANCE Lancas ter City Centr MAIN ENTR ANCE HEALTH INNOVATION CAMPUS ENTR SYMBOL KEY WALKS & ROUTES CAMPUS ZONES Accessible Parking co Colleges Library Shops/Refreshments Cycle Route North -

UNDERSTANDING the NEW NHS a Guide for Everyone Working and Training Within the NHS 2 Contents 3
England UNDERSTANDING THE NEW NHS A guide for everyone working and training within the NHS 2 Contents 3 NHS ENGLAND INFORMATION READER BOX Introduction 4 Directorate ◆ The NHS belongs to us all Medical Operations Patients and information Nursing Policy Commissioning development Foreword (Sir Bruce Keogh) 5 Finance Human resources NHS values 6 Publications Gateway Reference: 01486 ◆ NHS values and the NHS Constitution Document purpose Resources ◆ An overview of the Health and Social Care Act 2012 Document name Understanding The New NHS Structure of the NHS in England 8 Author NHS England ◆ The structure of the NHS in England Publication date 26 June 2014 ◆ Finance in the NHS: your questions answered Target audience Running the NHS 12 ◆ Commissioning in the NHS Additional circulation Clinicians working and training within the NHS, allied ◆ Delivering NHS services list health professionals, GPs ◆ Health and wellbeing in the NHS Description An updated guide on the structure and function of Monitoring the NHS 17 the NHS, taking into account the changes of the Health and Social Care Act 2012 ◆ Lessons learned and taking responsibility ◆ Regulation and monitoring in the NHS Action required None Contact details for Dr Felicity Taylor Working in the NHS 20 further information National Medical Director's Clinical ◆ Better training, better care Fellow, Medical Directorate Skipton House NHS leadership 21 80 London Road SE1 6LH ◆ Leading healthcare excellence www.england.nhs.uk/nhsguide/ Quality and innovation in the NHS 22 ◆ High-quality care for all Document status This is a controlled document. While this document may be printed, the The NHS in the United Kingdom 24 electronic version posted on the intranet is the controlled copy. -

Abstracts from the Medical Research Symposium for Students and Foundation Doctors 21 November 2018
Abstracts from the Medical Research Symposium for Students and Foundation Doctors 21 November 2018 November 2018 This abstract book has been produced using author-supplied copy and no editing has been undertaken. No responsibility is assumed for any claims, instructions, methods or drug dosages included in the abstracts: it is recommended that these are verified independently. Undergraduate Prizes Best undergraduate oral presentation Comparing Outcomes of Stent Retriever Thrombectomy versus Direct Aspiration for the Endovascular Treatment of Acute Anterior Circulation Stroke SE Thompson, R Jones, K Nader Birmingham Medical School Introduction Endovascular therapy with a stent retriever has been shown to be superior to standard medical therapy in the treatment of acute ischaemic stroke and is recommended by the American Stroke Association. Direct aspiration thrombectomy is another commonly used method increasing in popularity with advances in catheter technology. Few studies have directly compared the efficacy of the two methods, with current evidence equivocal. This study compares the stent retriever and direct aspiration methods. Patients and methods 101 patients presenting to the Queen Elizabeth Hospital Birmingham between January 2013-November 2017 with anterior circulation ischaemic stroke, fulfilling the criteria for endovascular therapy, were retrospectively analysed. 45 patients underwent first-line stent retriever thrombectomy and in 56 patients, direct aspiration was the primary method. The primary outcome was successful revascularization (TICI 2b/3) and secondary outcomes included procedural and clinical measures. Results Successful revascularization was achieved more frequently when direct aspiration was the primary chosen method (91.1% vs. 57.8%, p=0.000) and was significantly more likely to be successful with a single pass (p=0.005). -

Lancaster University Annual Report and Accounts 2011
LAncAsteR UniveRsity AnnUAL RepoRt And AccoUnts 2011 Lancaster University 1 1 Lancaster 0 2 LA1 4YW s t United Kingdom n u o c T: +44(0)1524 65201 c A www.lancs.ac.uk d n a ISBN 978-1-86220-290-0 t r o p e R y t i s r e v i n U r e t s a c n a L Lancaster University has been awarded the Carbon Trust Standard after taking action on climate change by reducing carbon emissions. The University has made an overall reduction of 245 tonnes of carbon or 0.9% averaged over the past three years. Lancaster University 1 Contents Vice-Chancellor’s review 2 Pro-Chancellor’s review 4 High notes of the year 6 A global university 12 Awards and distinctions 18 Advancing knowledge through research 30 Key facts and figures 42 Financial Statements 46 Operating and Financial Review for the year ended 31 July 2011 48 Statement of Corporate Governance 56 Independent Auditors’ Report to the Council of Lancaster University 61 Statement of Accounting Policies 62 Consolidated Income and Expenditure Account 64 Statement of Consolidated Historical Cost Surpluses and Deficits 65 Balance Sheets as at 31 July 20 11 66 Consolidated Cash Flow Statement 67 Reconciliation of Net Cash Flow to movement in Net Debt 68 Consolidated Statement of Total Recognised Gains and Losses 69 Notes to the Financial Statements 70 Chancellor’s Guild roll of honour 92 Chancellor’s Guild corporate donors and legacy pledges 93 Trusts, foundations and charities 94 Organisational chart 95 Lancaster University Report and Accounts 2011 3 ViCe-ChanCellor’s reView The growing reputation of Lancaster was marked this year by top ten placings in each of the UK’s major university been strengthened with a formal respond to this feedback to make sure The Government’s White Paper “Students collaboration with the Chinese Academy every department is outstanding. -

Annual Report and Accounts 2016/17
HC 172 Annual report and accounts 2016/17 Advise / Resolve / Learn NHS Resolution Annual report and accounts 2016/17 Presented to Parliament pursuant to Paragraph 6 of Schedule 15 of the National Health Service Act 2006. Ordered by the House of Commons to be printed 13 July 2017. This is a reproduction of the annual report and accounts laid before Parliament under the name of the NHS Litigation Authority - pages 5 to 165 remain unchanged HC 172 Contents © NHS Resolution 2017 (the operating name of the NHS Litigation Authority) The text in this document (this excludes, where present, the Royal Arms and all departmental or agency logos) may be reproduced free of charge in any format or medium provided that it is reproduced accurately and not in a misleading context. The material must be acknowledged as NHS Resolution copyright and the document title specified. Where third party material has been identified, permission from the respective copyright holder much be sought. Any enquiries related to this publication should be sent to us at 151 Buckingham Palace Road, London, SW1W 9SZ This publication is available at www.gov.uk/government/publications Print ISBN 9781474143813 Web ISBN 9781474143820 ID 18041725 07/17 Printed on paper containing 75% recycled fibre content minimum Printed in the UK by the Williams Lea Group on behalf of the Controller of Her Majesty’s Stationery Office Advise / Resolve / Learn Contents Contents Performance report 6 Chair’s welcome 8 Chief Executive’s report 10 Performance summary 12 What we do 12 Understanding our -

Health Research Authority Annual Report and Accounts, HC 312 Session 2012-2013
Health Research Authority Annual report and accounts HC 312 Health Research Authority Annual report and accounts for the period 1 December 2011* to 31 March 2012 Presented to Parliament pursuant to Section 232 (Schedule 15 paragraph 3) of the National Health Service Act 2006 Ordered by the House of Commons to be printed 5th July 2012 HC 312 LONDON: The Stationery Office £16.00 * The Health Research Authority (HRA) was established on 1 December 2011 The National Research Ethics Service (NRES) transferred to the HRA on 1 December 2011 Health Research Authority Annual Report and Accounts 2011-2012 © Health Research Authority (2012) The text of this document (this excludes, where present, the Royal Arms and all departmental and agency logos) may be reproduced free of charge in any format or medium providing that it is reproduced accurately and not in a misleading context The material must be acknowledged as Health Research Authority copyright and the document title specified. Where third party material has been identified, permission from the respective copyright holder must be sought. Any enquiries regarding this publication should be sent to us at HRA, Ground Floor, Skipton House, 80 London Road, London SE1 6LH This publication is also for download at www.official-documents.gov.uk This document is also available from our website at www.hra.nhs.uk ISBN: 9780102977561 Printed in the UK by The Stationery Office Limited on behalf of the Controller of Her Majesty’s Stationery Office ID: P002488734 07/12 Printed on paper containing 75% recycled fibre content minimum The Health Research Authority (HRA) was established on 1 December 2011. -

Becton Dickinson Blood Specimen Collection - Supply Disruption
Classification: Official Publication approval reference: B0933 To: All GPs in England. NHS England and NHS Improvement Pathology Incident Directors Skipton House Trust Chief Executives 80 London Road Trust EPRR leads London Trust Medical Directors SE1 6LH Trust Directors of Nursing Trust Procurement leads 26 August 2021 CCG Accountable Officers CCG EPRR leads CCG Medical leads CCG Procurement leads CCG Directors of Nursing ICS leads STP leads Dear Colleague, Becton Dickinson blood specimen collection - supply disruption Becton Dickinson has alerted us to a shortage of products from their Blood Specimen Collection Portfolio that is impacting across most main blood tube supplies. Working closely with DHSC, NHS Supply Chain, the MHRA and colleagues from the devolved administrations, given the impact on other parts of the UK, NHS England and NHS Improvement is co-ordinating the NHS’ response in England, on a national basis. The supply position remains constrained and is forecasted to become even more constrained over the coming weeks. While it is anticipated that the position will improve from the middle of September, overall supply is likely to remain challenging for a significant period. We have previously sent information in relation to this issue and will continue to provide updates as the supply situation develops. The tubes impacted by this shortage are: 1. 5mls Yellow top – SST 2 – clotted sample (e.g. U&E, LFTs etc) 2. Purple top – EDTA (e.g. FBC, HbA1c) Alternative products are being sought to alleviate these constraints; and all labs that will be switching to these supplies have received samples for testing and validation. However, it will take time for these products to be imported and delivered in volume to services.