Eric V. Goode, 2016 Behler Turtle Conservation Award Recipient
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Conservation Biology of Tortoises
The Conservation Biology of Tortoises Edited by Ian R. Swingland and Michael W. Klemens IUCN/SSC Tortoise and Freshwater Turtle Specialist Group and The Durrell Institute of Conservation and Ecology Occasional Papers of the IUCN Species Survival Commission (SSC) No. 5 IUCN—The World Conservation Union IUCN Species Survival Commission Role of the SSC 3. To cooperate with the World Conservation Monitoring Centre (WCMC) The Species Survival Commission (SSC) is IUCN's primary source of the in developing and evaluating a data base on the status of and trade in wild scientific and technical information required for the maintenance of biological flora and fauna, and to provide policy guidance to WCMC. diversity through the conservation of endangered and vulnerable species of 4. To provide advice, information, and expertise to the Secretariat of the fauna and flora, whilst recommending and promoting measures for their con- Convention on International Trade in Endangered Species of Wild Fauna servation, and for the management of other species of conservation concern. and Flora (CITES) and other international agreements affecting conser- Its objective is to mobilize action to prevent the extinction of species, sub- vation of species or biological diversity. species, and discrete populations of fauna and flora, thereby not only maintain- 5. To carry out specific tasks on behalf of the Union, including: ing biological diversity but improving the status of endangered and vulnerable species. • coordination of a programme of activities for the conservation of biological diversity within the framework of the IUCN Conserva- tion Programme. Objectives of the SSC • promotion of the maintenance of biological diversity by monitor- 1. -

Indian Freshwater Turtles, Which Are Usually Bigger
Fantastic Facts Indian Part 3 Freshwater Turtles Conservation / Threat Status of Turtles Many turtles, terrapins and tortoises are threatened with extinction, that is, dying out completely. Listed below are the turtles discussed in this article (from Part 1 to 3), with their status, or prospects of survival. Name Status (Global) Assam Roofed Turtle Endangered Cochin Forest Cane Turtle Endangered Crowned River Turtle Vulnerable Rock Terrapin Near Threatened Indian Flapshell Turtle Least Concern Indian Softshell Turtle Vulnerable Indian Narrow-headed Softshell Turtle Endangered Red-crowned Roofed Turtle Critically Endangered Northern River Terrapin Critically Endangered THE CATEGORIES Critically Endangered -- This is the highest category that a species can be assigned before “extinction”. It represents a “last ditch” effort to provide a warning to wildlife agencies and governments to activate management measures to protect the species before it disappears from the face of the earth. When a species is Critically Endangered, usually its chances of living for the next 100 years are very low. Often, its chances of surviving even for 10 years are not good at all ! Endangered -- This is the second highest threat category that a species can be assigned before it becomes further threatened e.g. Critically Endangered or Extinct. When a species is Endangered, its chances of survival as a species for the next 100 years are low. Vulnerable -- The IUCN Red List defines Vulnerable as when a species is not Critically Endangered or Endangered, but is still facing a high risk of extinction in the wild. This is the first threat category for ranking a species when it has some serious problems from human-related threats. -

Species Description Taxonomy Distribution
Gopher Tortoise Species Description Tortoises can be observed on all major landmasses in the world except for Australia and Antarctica. The Gopher Tortoise (Gopherus polyphemus) is one of four species that lives only in North America. It has a large dark-brown to grayish shell, elephantine limbs and a large grayish-black rounded head. Its shovel-like forefeet are used to excavate burrows that provide shelter (Ernst et al., 1994). The Gopher Tortoise can reach lengths of more than 35 centimeters and weigh over 10 kilograms, Photo: Pete Oxford and there are physical differences between males and females. Female tortoises have a flattened bottom shell, or plastron, and a small tail whereas males exhibit a highly concave plastron and large tail. Male Gopher Tortoises also have two visible scent glands beneath the chin (Ernst et al., 1994). Taxonomy The generic name of the Gopher Tortoise, Gopherus, is derived from the word gopher that was used to describe the burrowing habits of this species. The species name polyphemus refers to a cave-dwelling mythical creature (Ashton and Ashton, 2008). Daudin was the first to describe the Gopher Tortoise as polyphemus in 1802. Rafinesque used this species as the type or standard to define the features of Gopherus and assign this generic name to the North American tortoises (Schmidt, 1953). All four species in this genus exhibit adaptations for burrowing such as claws and flattened forelimbs. However, they can be distinguished based on anatomical features including limb-bone and skull structure (Ernst et al., 1994). The Gopher Tortoise is most closely related to the larger Mexican or Bolson Tortoise (G. -

Comparative Mitogenomics of Two Critically
Comparative Mitogenomics of Two Critically Endangered Turtles, Batagur Kachuga and Batagur Dhongoka (Testudines: Geoemydidae): Implications in Phylogenetics of Freshwater Turtles Ajit Kumar Wildlife Institute of India Prabhaker Yadav Wildlife Institute of India Aftab Usmani Wildlife Institute of India Syed Ainul Hussain Wildlife Institute of India Sandeep Kumar Gupta ( [email protected] ) Wildlife Institute of India Research Article Keywords: Mitochondrial genome, freshwater turtles, phylogenetic analysis, genetic relationship, evolutionary patterns Posted Date: July 13th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-690457/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/14 Abstract The Red-crowned roofed turtle (Batagur kachuga) and Three-striped roofed turtle (B. dhongoka) are ‘critically endangered’ turtles in the Geoemydidae family. Herein, we generated the novel mitochondrial genome sequence of B. kachuga (16,155) and B. dhongoka (15,620) and compared it with other turtles species. Batagur mitogenome has 22 transfer RNAs (tRNAs), 13 protein-coding genes (PCGs), two ribosomal RNAs (rRNAs), and one control region (CR). The genome composition was biased toward A + T, with positive AT-skew and negative GC-skew. In the examined species, all 13 PCGs were started by ATG codons, except COI gene, which was initiated by GTG. The majority of mito-genes were encoded on the heavy strand, except eight tRNAs and the ND6 region. We observed a typical cloverleaf structure for all tRNA, excluding tRNASer (AGN), where the base pairs of the dihydrouridine (DHU) arm were abridged. Bayesian Inference (BI) based phylogenetic analysis was constructed among 39 species from six Testudines families, exhibited a close genetic relationship between Batagur and Pangshura with a high supporting value (PP ~ 0.99). -

Phylogenetic Relationships Within the Batagur Complex (Testudines: Emydidae: Batagurinae) Jean M
Eastern Illinois University The Keep Masters Theses Student Theses & Publications 1993 Phylogenetic Relationships Within the Batagur Complex (Testudines: Emydidae: Batagurinae) Jean M. Capler This research is a product of the graduate program in Zoology at Eastern Illinois University. Find out more about the program. Recommended Citation Capler, Jean M., "Phylogenetic Relationships Within the Batagur Complex (Testudines: Emydidae: Batagurinae)" (1993). Masters Theses. 2114. https://thekeep.eiu.edu/theses/2114 This is brought to you for free and open access by the Student Theses & Publications at The Keep. It has been accepted for inclusion in Masters Theses by an authorized administrator of The Keep. For more information, please contact [email protected]. THESIS REPRODUCTION CERTIFICATE TO: Graduate Degree Candidat.es who have written formal theses. SUBJECT: Permi~sion to reproduce theses. The University Library is r~c;:eiving a number of requests from other institutions asklng permission to reproduce dissertations for inclusion in thelr library holdings. Although no copyr~ght laws are involved, we feel that professional courtesy demands that permission be obtained from the author before we allow theses to be copied. Please sign one of the following statements: Booth Library of Eastern Illinois University has my permission to lend my thesis to a reputable college or university for the purpose of copying it for inclusion in that institution's library or research holdings. Date I respectfully request Booth Library of Easter,n Illinois University not ~llow my thesis be reproduced because ---~~~~--~~~~~---........ Date Author m L Phylogenetic Relationships Within The Batagur Complex (Testudines: Emydidae: Batagurinae) (TITLE) BY Jean M. Capler THESIS SUBMITIED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master Of Science IN THE GRADUATE SCHOOL, EASTERN ILLINOIS UNIVERSITY CHARLESTON, ILLINOIS 1993 YEAR I HEREBY RECOMMEND THIS THESIS BE ACCEPTED AS FULFILLING 17 ~ \'\93 DA . -

15Th Annual Symposium on the Conservation and Biology of Tortoises and Freshwater Turtles
CHARLESTON, SOUTH CAROLINA 2017 15th Annual Symposium on the Conservation and Biology of Tortoises and Freshwater Turtles Joint Annual Meeting of the Turtle Survival Alliance and IUCN Tortoise & Freshwater Turtle Specialist Group Program and Abstracts August 7 - 9 2017 Charleston, SC Additional Conference Support Provided by: Kristin Berry, Herpetologiccal Review, John Iverson, Robert Krause,George Meyer, David Shapiro, Anders Rhodin, Brett and Nancy Stearns, and Reid Taylor Funding for the 2016 Behler Turtle Conservation Award Provided by: Brett and Nancy Stearns, Chelonian Research Foundation, Deb Behler, George Meyer, IUCN Tortoise and Freshwater Turtle Specialist Group, Leigh Ann and Matt Frankel and the Turtle Survival Alliance TSA PROJECTS TURTLE SURVIVAL ALLIANCE 2017 Conference Highlights In October 2016, the TSA opened the Keynote: Russell Mittermeier Tortoise Conservation Center in southern Madagascar that will provide long- Priorities and Opportunities in Biodiversity Conservation term care for the burgeoning number of tortoises seized from the illegal trade. Russell A. Mittermeier is The TSA manages over 7,800 Radiated Executive Vice Chair at Con- Tortoises in seven rescue facilities. servation International. He served as President of Conser- vation International from 1989 to 2014. Named a “Hero for the Planet” by TIME magazine, Mittermeier is regarded as a world leader in the field of biodiversity and tropical forest conservation. Trained as a primatologist and herpetologist, he has traveled widely in over 160 countries on seven continents, and has conducted field work in more than 30 − focusing particularly on Amazonia (especially Brazil The TSA-Myanmar team and our vet- and Suriname), the Atlantic forest region of Brazil, and Madagascar. -
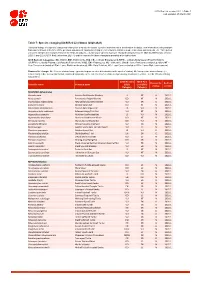
Table 7: Species Changing IUCN Red List Status (2020-2021)
IUCN Red List version 2021-1: Table 7 Last Updated: 25 March 2021 Table 7: Species changing IUCN Red List Status (2020-2021) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2020 (IUCN Red List version 2020-3) and 2021 (IUCN Red List version 2021-1) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2020) List (2021) change version Category -

TSA Magazine 2008
TSATURTLE SURVIVAL ALLIANCE AUGUST 2008 An IUCN Partnership Network for Sustainable Captive Management of Freshwater Turtles & Tortoises — www.TurtleSurvival.org N O MANS L A H K ITC M GIANT YANGTZE SOFTSHELL TURTLE, RAFETUS SWINHOEI (SEE ARTICLE P. 4) 1 From the TSA Co-Chairs As you read this eighth edition of the TSA newsletter, reflect back on how far this publication has come since 2001. It’s difficult to continue to call this a newsletter. Perhaps TSA magazine or annual report would be a better name. Regardless, we hope you like the new polished format and appreciate the extra pages. Putting this publication together takes more and more effort every year, and that is certainly a positive reflection on the growth of our organization. We have a lot going on around the globe, and our reputation for doing good turtle conservation work continues to grow. The TSA is becoming well known for taking decisive conservation action and being unafraid to take risks when situ- ations warrant. There can be no better example of this than our top story for 2008--the historic attempt to breed the last two Yangtze giant softshell turtles, Rafetus swinhoei, in China. Under the able leadership of Dr.Gerald Kuchling, and with superb support and assistance from Lu Shunquing of WCS-China, an agreement was reached to unite the only known living female Rafetus at Changsha Zoo with an ancient male at Suzhou Zoo. At least three workshops were held to reach this agreement. But once this happened, TSA began to raise funds in anticipation of an event we knew would be expensive, high profile, and risky. -

Setting the Stage for Understanding Globalization of the Asian Turtle Trade
Setting the Stage for Understanding Globalization of the Asian Turtle Trade: Global, Asian, and American Turtle Diversity, Richness, Endemism, and IUCN Red List Threat Levels Anders G.J. Rhodin and Peter Paul van Dijk IUCN Tortoise and Freshwater Turtle Specialist Group, Chelonian Research Foundation, Conservation International Thursday, January 20, 2011 New Species Described 2010 Photo C. Hagen Graptemys pearlensis - Pearl River Map Turtle Louisiana and Mississippi, USA Red List: Not Evaluated [Endangered] Thursday, January 20, 2011 IUCN/SSC Tortoise and Freshwater Turtle Specialist Group Founded 1980 www.iucn-tftsg.org Thursday, January 20, 2011 International Union for the Conservation of Nature / Species Survival Commission www.iucn.org Thursday, January 20, 2011 Convention on International Trade in Endangered Species of Fauna and Flora www.cites.org Thursday, January 20, 2011 Chelonian Conservation and Biology Thomson Reuters’ ISI Journal Citation Impact Factor currently ranks CCB among the top 100 zoology journals worldwide www.chelonianjournals.org Thursday, January 20, 2011 Conservation Biology of Freshwater Turtles and Tortoises www.iucn-tftsg.org/cbftt Thursday, January 20, 2011 IUCN Tortoise and Freshwater Turtle Specialist Group Members: Work or Focus - 2010 274 Members - 107 Countries Thursday, January 20, 2011 Species, Additional Subspecies, and Total Taxa of Turtles and Tortoises 500 Species Add. Subspecies 375 Total Taxa 250 125 0 1758176617831789179218011812183518441856187318891909193419551961196719771979198619891992199420062007200820092010 Currently Recognized: 334 species, 127 add. subspecies, 461 total taxa Thursday, January 20, 2011 Tortoise and Freshwater Turtle Species Richness Buhlmann, Akre, Iverson, Karapatakis, Mittermeier, Georges, Rhodin, van Dijk, and Gibbons. 2009. Chelonian Conservation and Biology 8:116–149. Thursday, January 20, 2011 Tortoise and Freshwater Turtle Species Richness – Global Rankings 1. -

TCF Summary Activity Report 2002–2018
Turtle Conservation Fund • Summary Activity Report 2002–2018 Turtle Conservation Fund A Partnership Coalition of Leading Turtle Conservation Organizations and Individuals Summary Activity Report 2002–2018 1 Turtle Conservation Fund • Summary Activity Report 2002–2018 Recommended Citation: Turtle Conservation Fund [Rhodin, A.G.J., Quinn, H.R., Goode, E.V., Hudson, R., Mittermeier, R.A., and van Dijk, P.P.]. 2019. Turtle Conservation Fund: A Partnership Coalition of Leading Turtle Conservation Organi- zations and Individuals—Summary Activity Report 2002–2018. Lunenburg, MA and Ojai, CA: Chelonian Research Foundation and Turtle Conservancy, 54 pp. Front Cover Photo: Radiated Tortoise, Astrochelys radiata, Cap Sainte Marie Special Reserve, southern Madagascar. Photo by Anders G.J. Rhodin. Back Cover Photo: Yangtze Giant Softshell Turtle, Rafetus swinhoei, Dong Mo Lake, Hanoi, Vietnam. Photo by Timothy E.M. McCormack. Printed by Inkspot Press, Bennington, VT 05201 USA. Hardcopy available from Chelonian Research Foundation, 564 Chittenden Dr., Arlington, VT 05250 USA. Downloadable pdf copy available at www.turtleconservationfund.org 2 Turtle Conservation Fund • Summary Activity Report 2002–2018 Turtle Conservation Fund A Partnership Coalition of Leading Turtle Conservation Organizations and Individuals Summary Activity Report 2002–2018 by Anders G.J. Rhodin, Hugh R. Quinn, Eric V. Goode, Rick Hudson, Russell A. Mittermeier, and Peter Paul van Dijk Strategic Action Planning and Funding Support for Conservation of Threatened Tortoises and Freshwater -

Habitat Relations of Juvenile Gopher Tortoises and a Preliminary Report Of
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1-1-1993 Habitat relations of juvenile gopher tortoises and a preliminary report of Upper Respiratory Tract Disease (URTD) in gopher tortoises Stephen Michael Beyer Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Recommended Citation Beyer, Stephen Michael, "Habitat relations of juvenile gopher tortoises and a preliminary report of Upper Respiratory Tract Disease (URTD) in gopher tortoises" (1993). Retrospective Theses and Dissertations. 18016. https://lib.dr.iastate.edu/rtd/18016 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Habitat relations of juvenile gopher tortoises and a preliminary report of Upper Respiratory Tract Disease (URTD) in gopher tortoises by Stephen Michael Beyer A Thesis Submitted to the Graduate Faculty in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE Department: Animal Ecology Interdepartmental Major: Ecology and Evolutionary Biology Signatures have been redacted for privacy Iowa State University Ames, Iowa 1993 ii TABLE OF CONTENTS GENERAL INTRODUCTION 1 PAPER I HOME RANGE, ACTIVITY, AND HABITAT SELECTION OF JUVENILE GOPHER -

TSA Magazine 2015
A PUBLICATION OF THE TURTLE SURVIVAL ALLIANCE Turtle Survival 2015 RICK HUDSON FROM THE PRESIDENT’S DESK TSA’s Commitment to Zero Turtle Extinctions more than just a slogan Though an onerous task, this evaluation process is completely necessary if we are to systematically work through the many spe- cies that require conservation actions for their survival. Determining TSA’s role for each species is important for long-term planning and the budgeting process, and to help us identify areas around the globe where we need to develop new field programs. In Asia for example, Indonesia and Vietnam, with nine targeted species each, both emerged as high priority countries where we should be working. Concurrently, the Animal Management plan identified 32 species for man- agement at the Turtle Survival Center, and the associated space requirements imply a signifi- cant investment in new facilities. Both the Field Conservation and Animal Management Plans provide a blueprint for future growth for the TSA, and document our long-term commitment. Failure is not an option for us, and it will require a significant investment in capital and expansion if we are to make good on our mission. As if to test TSA’s resolve to make good on our commitment, on June 17 the turtle conser- vation community awoke to a nightmare when we learned of the confiscation of 3,800 Palawan Forest Turtles in the Philippines. We dropped everything and swung into action and for weeks to come, this crisis and the coordinated response dominated our agenda. In a show of PHOTO CREDIT: KALYAR PLATT strength and unity, turtle conservation groups from around the world responded, deploying Committed to Zero Turtle Extinctions: these species that we know to be under imminent both staff and resources.